7RUU
 
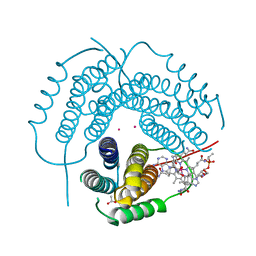 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
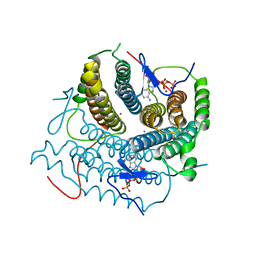 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
2RNA
 
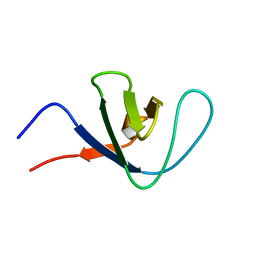 | | Itk SH3 average minimized | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Severin, A.J. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: murine Itk SH3 domain
To be Published
|
|
5HXC
 
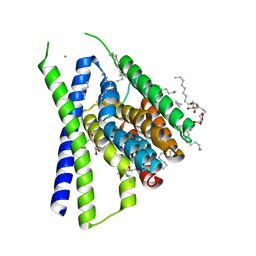 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 20 mM Na+ and zero Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CALCIUM ION, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-01-30 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger
Nat.Struct.Mol.Biol., 23, 2016
|
|
7RUV
 
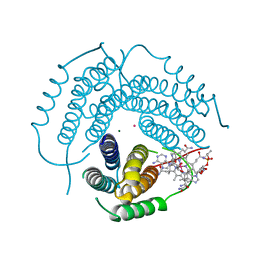 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
2RND
 
 | |
6XS6
 
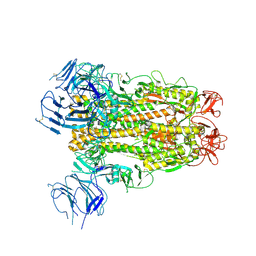 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
2RNJ
 
 | | NMR Structure of The S. Aureus VraR DNA Binding Domain | | Descriptor: | Response regulator protein vraR | | Authors: | Donaldson, L.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the Staphylococcus aureus Response Regulator VraR DNA Binding Domain Reveals a Dynamic Relationship between It and Its Associated Receiver Domain
Biochemistry, 47, 2008
|
|
5HW7
 
 | |
2RI5
 
 | |
7SEX
 
 | | M. tb EgtD in complex with TGX221 | | Descriptor: | 7-methyl-2-morpholin-4-yl-9-[(1~{R})-1-phenylazanylethyl]-3~{H}-pyrido[1,2-a]pyrimidin-4-one, GLYCEROL, Histidine N-alpha-methyltransferase | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-10-02 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Mycobacterium tuberculosis EgtD target both substrate binding sites to limit hercynine production.
Sci Rep, 11, 2021
|
|
2RNQ
 
 | |
2G97
 
 | |
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
7D4S
 
 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
5HXA
 
 | |
2ROG
 
 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
5HZ6
 
 | |
7D4O
 
 | | cyclic trinucleotide synthase CdnD in complex with ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
2GHD
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
6XJF
 
 | |
2R6R
 
 | | Aquifex aeolicus FtsZ | | Descriptor: | Cell division protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Trambaiolo, D, Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-06 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the conformational variability of FtsZ.
J.Mol.Biol., 373, 2007
|
|
2GHY
 
 | | Novel Crystal Form of the ColE1 Rom Protein | | Descriptor: | Regulatory protein rop | | Authors: | Jang, S.B, Jeong, M.S, Carter, R.J, Holbrook, E.L, Comolli, L.R, Holbrook, S.R. | | Deposit date: | 2006-03-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel crystal form of the ColE1 Rom protein.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6XXE
 
 | |
7SEY
 
 | | M. tb EgtD in complex with SGH | | Descriptor: | 2-amino-1-[(3S)-3-methyl-4-(4-methylisoquinoline-5-sulfonyl)-1,4-diazepan-1-yl]ethan-1-one, Histidine N-alpha-methyltransferase | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-10-02 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Inhibitors of Mycobacterium tuberculosis EgtD target both substrate binding sites to limit hercynine production.
Sci Rep, 11, 2021
|
|
