1JJT
 
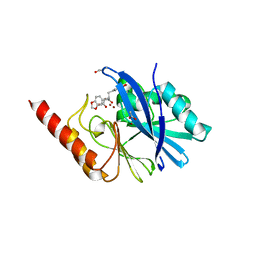 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A BIARYL SUCCINIC ACID INHIBITOR (1) | | Descriptor: | 2,3-BIS-BENZO[1,3]DIOXOL-5-YLMETHYL-SUCCINIC ACID, ACETATE ION, IMP-1 METALLO BETA-LACTAMASE, ... | | Authors: | Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-beta-lactamase.
J.Biol.Chem., 276, 2001
|
|
1JJU
 
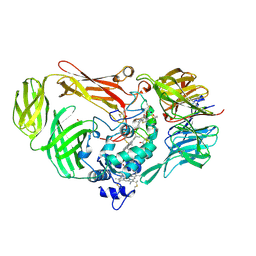 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JJV
 
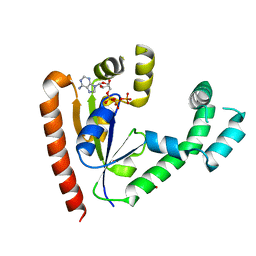 | | DEPHOSPHO-COA KINASE IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEPHOSPHO-COA KINASE, MERCURY (II) ION, ... | | Authors: | Obmolova, G, Teplyakov, A, Bonander, N, Eisenstein, E, Howard, A.J, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae.
J.Struct.Biol., 136, 2001
|
|
1JJW
 
 | |
1JJX
 
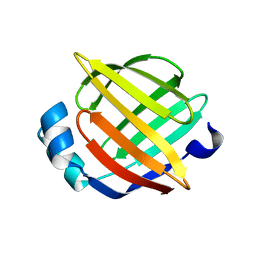 | | Solution Structure of Recombinant Human Brain-type Fatty acid Binding Protein | | Descriptor: | BRAIN-TYPE FATTY ACID BINDING PROTEIN | | Authors: | Rademacher, M, Zimmerman, A.W, Rueterjans, H, Veerkamp, J.H, Luecke, C. | | Deposit date: | 2001-07-10 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of fatty acid-binding protein from human brain.
Mol.Cell.Biochem., 239, 2002
|
|
1JJZ
 
 | |
1JK0
 
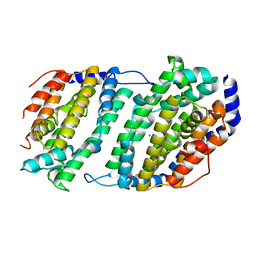 | | Ribonucleotide reductase Y2Y4 heterodimer | | Descriptor: | ZINC ION, ribonucleoside-diphosphate reductase small chain 1, ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2001-07-10 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JK1
 
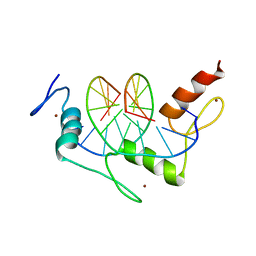 | | Zif268 D20A Mutant Bound to WT DNA Site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*G)-3', 5'-D(*TP*CP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1JK2
 
 | | Zif268 D20A mutant bound to the GCT DNA site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1JK3
 
 | | Crystal structure of human MMP-12 (Macrophage Elastase) at true atomic resolution | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MACROPHAGE METALLOELASTASE, ... | | Authors: | Lang, R, Kocourek, A, Braun, M, Tschesche, H, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 A crystal structure.
J.Mol.Biol., 312, 2001
|
|
1JK4
 
 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
1JK6
 
 | | UNCOMPLEXED DES 1-6 BOVINE NEUROPHYSIN | | Descriptor: | NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2003
|
|
1JK7
 
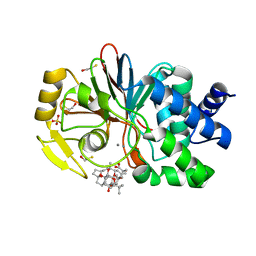 | | CRYSTAL STRUCTURE OF THE TUMOR-PROMOTER OKADAIC ACID BOUND TO PROTEIN PHOSPHATASE-1 | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Bateman, K.S, Cherney, M.M, Das, A.K, James, M.N. | | Deposit date: | 2001-07-11 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.
J.Biol.Chem., 276, 2001
|
|
1JK8
 
 | |
1JK9
 
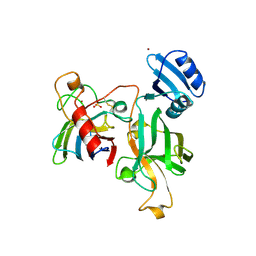 | | Heterodimer between H48F-ySOD1 and yCCS | | Descriptor: | SULFATE ION, ZINC ION, copper chaperone for superoxide dismutase, ... | | Authors: | Lamb, A.L, Torres, A.S, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Heterodimeric structure of superoxide dismutase in complex with its metallochaperone.
Nat.Struct.Biol., 8, 2001
|
|
1JKA
 
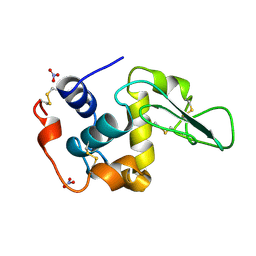 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ASP | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKB
 
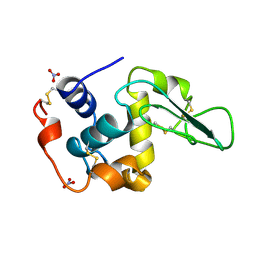 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKC
 
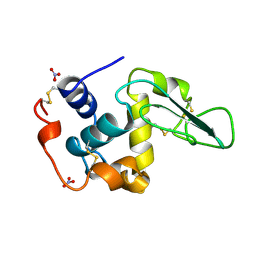 | | HUMAN LYSOZYME MUTANT WITH TRP 109 REPLACED BY PHE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKD
 
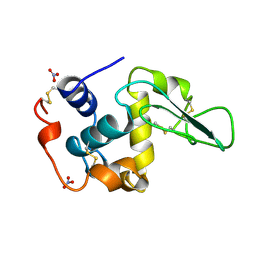 | | HUMAN LYSOZYME MUTANT WITH TRP 109 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKE
 
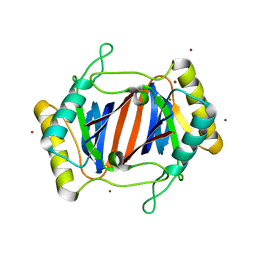 | | D-Tyr tRNATyr deacylase from Escherichia coli | | Descriptor: | D-Tyr-tRNATyr deacylase, ZINC ION | | Authors: | Ferri-Fioni, M.L, Schmitt, E, Soutourina, J, Plateau, P, Mechulam, Y, Blanquet, S. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases.
J.Biol.Chem., 276, 2001
|
|
1JKF
 
 | | Holo 1L-myo-inositol-1-phosphate Synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, myo-inositol-1-phosphate synthase | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1JKG
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA nuclear export factor | | Descriptor: | TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
1JKH
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JKI
 
 | | myo-Inositol-1-phosphate Synthase Complexed with an Inhibitor, 2-deoxy-glucitol-6-phosphate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-DEOXY-GLUCITOL-6-PHOSPHATE, AMMONIUM ION, ... | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1JKJ
 
 | | E. coli SCS | | Descriptor: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase.
Biochemistry, 41, 2002
|
|
