4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
3BN1
 
 | | Crystal structure of GDP-perosamine synthase | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, Perosamine synthetase, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GDP-Perosamine Synthase: Structural Analysis and Production of a Novel Trideoxysugar
Biochemistry, 47, 2008
|
|
1ZXF
 
 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1YQP
 
 | | T268N mutant cytochrome domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
1UPW
 
 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
3EFD
 
 | | The crystal structure of the cytoplasmic domain of KcsA | | Descriptor: | FabH, FabL, KcsA | | Authors: | Uysal, S, Vasquez, V, Tereshko, V, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DR7
 
 | | GDP-perosamine synthase from Caulobacter crescentus with bound GDP-3-deoxyperosamine | | Descriptor: | (2R,3S,5S,6R)-5-amino-3-hydroxy-6-methyl-oxan-2-yl, 1,2-ETHANEDIOL, Putative perosamine synthetase | | Authors: | Holden, H.M, Cook, P.D, Carney, A.E. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of GDP-linked sugars in the active site of GDP-perosamine synthase
Biochemistry, 47, 2008
|
|
3DXL
 
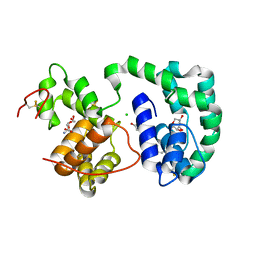 | | Crystal structure of AeD7 from Aedes Aegypti | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Allergen Aed a 2, CHLORIDE ION, ... | | Authors: | Andersen, J.F, Calvo, E, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2008-07-24 | | Release date: | 2009-02-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DY9
 
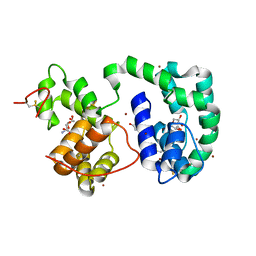 | | Crystal structure of AeD7 potassium bromide soak | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, D7 protein, ... | | Authors: | Andersen, J.F, Calvo, E, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2008-07-25 | | Release date: | 2009-02-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1YQO
 
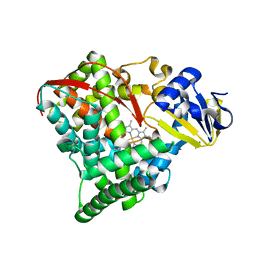 | | T268A mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
3DJQ
 
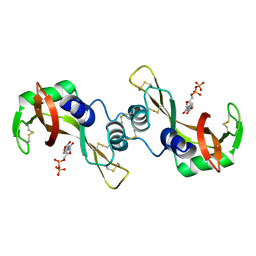 | | Bovine Seminal Ribonuclease- Uridine 5' diphosphate complex | | Descriptor: | Seminal ribonuclease, URIDINE-5'-DIPHOSPHATE | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3DJV
 
 | | Bovine Seminal Ribonuclease- cytidine 3' phosphate complex | | Descriptor: | CYTIDINE-3'-MONOPHOSPHATE, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3DR4
 
 | | GDP-perosamine synthase K186A mutant from Caulobacter crescentus with bound sugar ligand | | Descriptor: | 1,2-ETHANEDIOL, Putative perosamine synthetase, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S,5S,6R)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-6-methyltetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Holden, H.M, Cook, P.D, Carney, A.E. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of GDP-linked sugars in the active site of GDP-perosamine synthase
Biochemistry, 47, 2008
|
|
5AW7
 
 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 11.3 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVQ
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 0.75 min. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW1
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 85 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
2ZAU
 
 | | Crystal structure of an N-terminally truncated selenophosphate synthetase from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Selenide, water dikinase | | Authors: | Sekine, S, Matsumoto, E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an N-terminally truncated selenophosphate synthetase from Aquifex aeolicus
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2A6P
 
 | | Structure Solution to 2.2 Angstrom and Functional Characterisation of the Open Reading Frame Rv3214 from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, POSSIBLE PHOSPHOGLYCERATE MUTASE GPM2, SULFATE ION | | Authors: | Watkins, H.A, Yu, M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-03 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of Rv3214 from Mycobacterium tuberculosis, a Protein with Conflicting Functional Annotations, Leads to Its Characterization as a Phosphatase.
J.Bacteriol., 188, 2006
|
|
3Q3Q
 
 | |
5AVS
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 3.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW4
 
 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 1.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVT
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW3
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 100 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
4ZI9
 
 | | Structure of mouse clustered PcdhgA1 EC1-3 | | Descriptor: | CALCIUM ION, MCG133388, isoform CRA_t | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
