5K7K
 
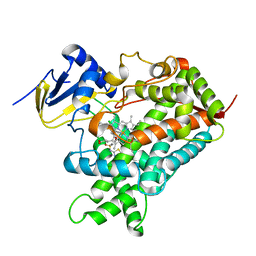 | |
8HOU
 
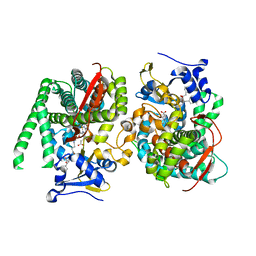 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Im-N-C4-Phe-Phe | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-N-C4-Phe-Phe, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOR
 
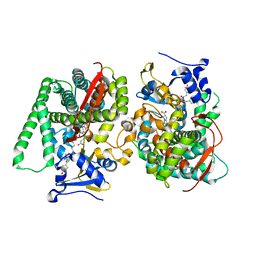 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CH3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CH3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOS
 
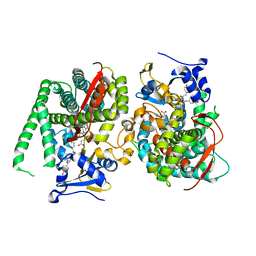 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4NO2)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4NO2)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOP
 
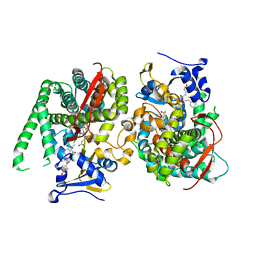 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HON
 
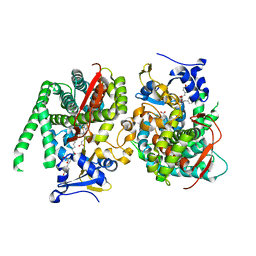 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HOQ
 
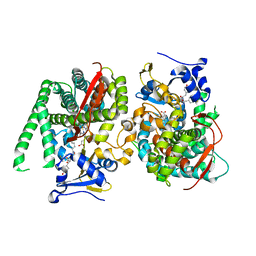 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CF3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CF3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6TO2
 
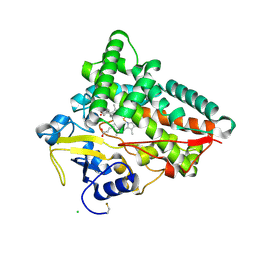 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5YSW
 
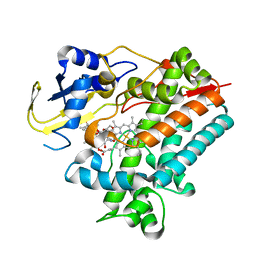 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5G
 
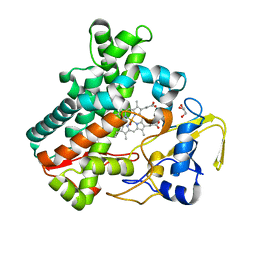 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5K
 
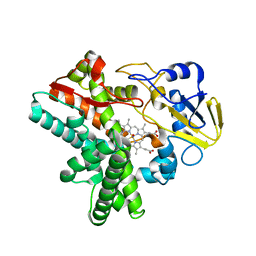 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6A17
 
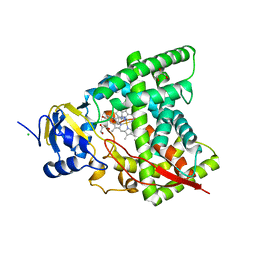 | | Crystal structure of CYP90B1 in complex with brassinazole | | Descriptor: | (2R,3S)-4-(4-chlorophenyl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)butan-2-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A15
 
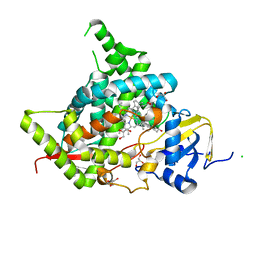 | | Structure of CYP90B1 in complex with cholesterol | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A18
 
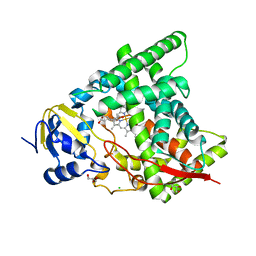 | | Crystal structure of CYP90B1 in complex with 1,6-hexandiol | | Descriptor: | CHLORIDE ION, Cytochrome P450 90B1, GLYCEROL, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A16
 
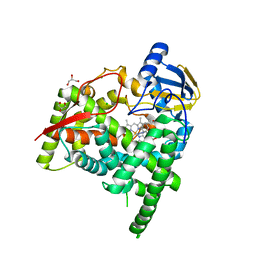 | | Crystal structure of CYP90B1 in complex with uniconazole | | Descriptor: | (1E,3S)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)pent-1-en-3-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
1ODO
 
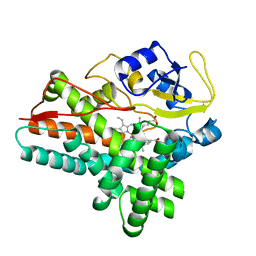 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
4BF4
 
 | |
3ZPI
 
 | | PikC D50N mutant in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 HYDROXYLASE PIKC, FORMIC ACID, ... | | Authors: | Podust, L.M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
5WBG
 
 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | Descriptor: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
5E0E
 
 | |
5E58
 
 | | Crystal Structure Of Cytochrome P450 2B35 from Desert Woodrat Neotoma Lepida in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 family 2 subfamily B, ... | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Function Analysis of Mammalian CYP2B Enzymes Using 7-Substituted Coumarin Derivatives as Probes: Utility of Crystal Structures and Molecular Modeling in Understanding Xenobiotic Metabolism.
Mol.Pharmacol., 89, 2016
|
|
5LIV
 
 | | Crystal structure of myxobacterial CYP260A1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 CYP260A1,Cytochrome P450 CYP260A1, DIMETHYL SULFOXIDE, ... | | Authors: | Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of CYP260A1 from Sorangium cellulosum to investigate the 1 alpha-hydroxylation of a mineralocorticoid.
FEBS Lett., 590, 2016
|
|
