1QGU
 
 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, DITHIONITE-REDUCED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-06 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
4IFA
 
 | | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Shatsman, S, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames
To be Published
|
|
3VNT
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with a [1,3]thiazolo[5,4-b]pyridine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H. | | Deposit date: | 2012-01-17 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds
J.Med.Chem., 55, 2012
|
|
3VO3
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with imidazo[1,2-b]pyridazine derivative | | Descriptor: | 1,2-ETHANEDIOL, N-[3-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)phenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H, Okada, K. | | Deposit date: | 2012-01-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-[5-({2-[(cyclopropylcarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}oxy)-2-methylphenyl]-1,3-dimethyl-1H-pyrazole-5-carboxamide (TAK-593), a highly potent VEGFR2 kinase inhibitor
Bioorg.Med.Chem., 21, 2013
|
|
1VDV
 
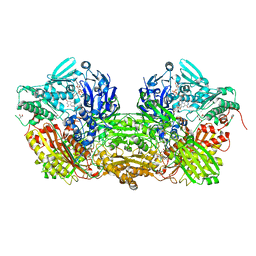 | | Bovine Milk Xanthine Dehydrogenase Y-700 Bound Form | | Descriptor: | 1-[3-CYANO-4-(NEOPENTYLOXY)PHENYL]-1H-PYRAZOLE-4-CARBOXYLIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Fukunari, A, Okamoto, K, Nishino, T, Eger, B.T, Pai, E.F, Kamezawa, M, Yamada, I, Kato, N. | | Deposit date: | 2004-03-25 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Y-700 [1-[3-Cyano-4-(2,2-dimethylpropoxy)phenyl]-1H-pyrazole-4-carboxylic acid]: a potent xanthine oxidoreductase inhibitor with hepatic excretion
J.Pharmacol.Exp.Ther., 311, 2004
|
|
3RJO
 
 | |
5TW9
 
 | | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Iron uptake system component EfeO | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-11 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis.
To Be Published
|
|
4KTT
 
 | | Structural insights of MAT enzymes: MATa2b complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine adenosyltransferase 2 subunit beta, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
3TBJ
 
 | | The 1.7A crystal structure of Actibind a T2 ribonucleases as antitumorigenic agents | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Almog, O, Gonzalez, A. | | Deposit date: | 2011-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of ACTIBIND suggests a mode of action for T2 ribonucleases as antitumorigenic agents.
J.Med.Chem., 55, 2012
|
|
1K5H
 
 | | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | | Authors: | Reuter, K, Sanderbrand, S, Jomaa, H, Wiesner, J, Steinbrecher, I, Beck, E, Hintz, M, Klebe, G, Stubbs, M.T. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1QH1
 
 | | NITROGENASE MOFE PROTEIN FROM KLEBSIELLA PNEUMONIAE, PHENOSAFRANIN OXIDIZED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-10 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
3AY6
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 A258F mutant in complex with NADH and D-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Glucose 1-dehydrogenase 4, ... | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
1KV9
 
 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
2RCC
 
 | |
7LO2
 
 | | S-adenosylmethionine synthetase cocrystallized with CTP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
1QH8
 
 | | NITROGENASE MOFE PROTEIN FROM KLEBSIELLA PNEUMONIAE, AS-CRYSTALLIZED (MIXED OXIDATION) STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
1R0K
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ACETATE ION | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1I30
 
 | | E. Coli Enoyl Reductase +NAD+SB385826 | | Descriptor: | 1,3,4,9-TETRAHYDRO-2-(HYDROXYBENZOYL)-9-[(4-HYDROXYPHENYL)METHYL]-6-METHOXY-2H-PYRIDO[3,4-B]INDOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seefeld, M.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-04-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
3UWQ
 
 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
3VBW
 
 | |
3W87
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[5-(methoxycarbonyl)naphthalen-2-yl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103
To be Published
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
1THG
 
 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
4KTV
 
 | | Structural insights of MAT enzymes: MATa2b complexed with adenosine and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
