7AIJ
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
5YLF
 
 | | MCR-1 complex with D-glucose | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
6ORP
 
 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab897NHP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab897NHP antibody Fab heavy chain, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
6SUD
 
 | | Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Reducing-end xylose-releasing exo-oligoxylanase Rex8A, ... | | Authors: | Jimenez-Ortega, E, Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-09-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Febs J., 287, 2020
|
|
7S3I
 
 | | Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 2) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Piper, S.J, Danev, R. | | Deposit date: | 2021-09-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural and functional diversity among agonist-bound states of the GLP-1 receptor.
Nat.Chem.Biol., 18, 2022
|
|
6ZBH
 
 | |
5I5V
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.94A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIENOPYRIMIDINE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxylic acid, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
6ZBC
 
 | |
6ZBG
 
 | |
6ZBJ
 
 | |
9NBG
 
 | | H-1 Parvovirus VLP - Glycan [s(Lex)2] | | Descriptor: | Major capsid protein, N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Busuttil, K.B, Bennett, A.B, McKenna, R. | | Deposit date: | 2025-02-13 | | Release date: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Mapping the Sialic Acid Binding Sites of LuIII and H-1 Parvovirus
To Be Published
|
|
5NX5
 
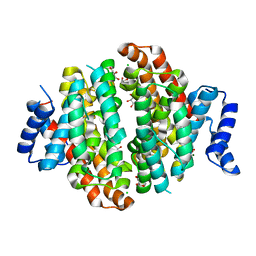 | | Crystal structure of Linalool/Nerolidol synthase from Streptomyces clavuligerus in complex with 2-fluorogeranyl diphosphate | | Descriptor: | (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
8RCL
 
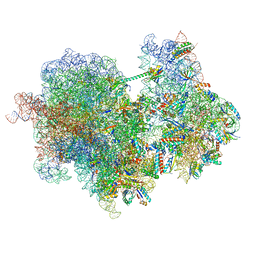 | |
8BSM
 
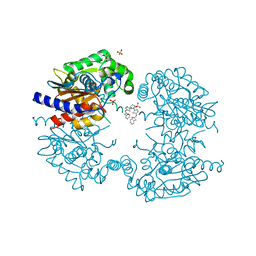 | | Human GLS in complex with compound 18 | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.782 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
5YR6
 
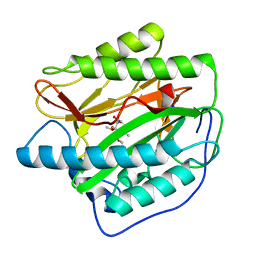 | | Human methionine aminopeptidase type 1b (F309L mutant) in complex with TNP470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Pillalamarri, V, Arya, T, Addlagatta, A. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of natural product ovalicin sensitive type 1 methionine aminopeptidases: molecular and structural basis.
Biochem. J., 476, 2019
|
|
5O19
 
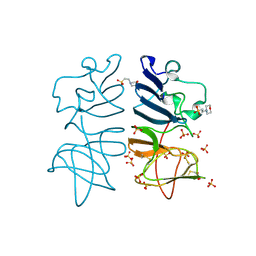 | |
8FS5
 
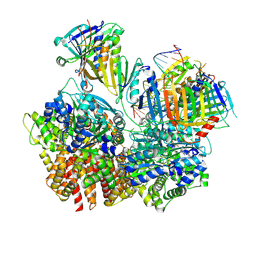 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS4
 
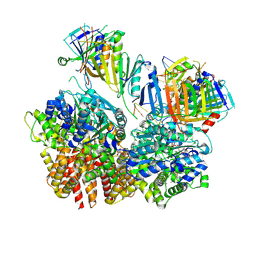 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS3
 
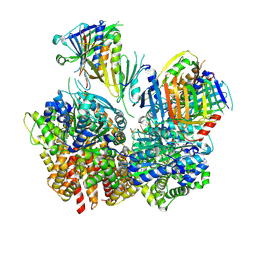 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8VB1
 
 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
8FS8
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
6GTB
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with FB211 | | Descriptor: | 1,2-ETHANEDIOL, 17-beta-hydroxysteroid dehydrogenase 14, 3-[6-(3-hydroxyphenyl)pyridin-2-yl]benzoic acid, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2018-06-18 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
7BZI
 
 | |
