6POL
 
 | | Crystal structure of the human NELL1 EGF1-3-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
1JZ3
 
 | |
6CN9
 
 | | Crystal structure of the Kinase domain of WNK1 | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J. | | Deposit date: | 2018-03-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension.
Structure, 12, 2004
|
|
8RUS
 
 | | Hen egg-white lysozyme (HEWL) structure from EuXFEL FXE, multi-hit Droplet-on-Demand (DoD) injection, 9.3 keV photon energy, space group P432121 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perrett, S, van Thor, J.J. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE.
Struct Dyn., 11, 2024
|
|
1JZ6
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTO-TETRAZOLE | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Juers, D.H, Heightman, T.D, Vasella, A, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
3KDS
 
 | | apo-FtsH crystal structure | | Descriptor: | Cell division protein FtsH, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-naphthalen-2-yl-L-alanyl-L-alaninamide, ZINC ION | | Authors: | Bieniossek, C, Niederhauser, B, Baumann, U. | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2J9T
 
 | | BipD of Burkholderia Pseudomallei | | Descriptor: | BORIC ACID, CITRATE ANION, MEMBRANE ANTIGEN | | Authors: | Johnson, S, Roversi, P, Field, T, Deane, J.E, Galyov, E, Lea, S.M. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
6PL4
 
 | | TRK-A IN COMPLEX WITH LIGAND 1 | | Descriptor: | High affinity nerve growth factor receptor, N-{[5-(methoxymethyl)-2-(trifluoromethoxy)phenyl]methyl}-N'-(8-methyl-2-phenylimidazo[1,2-a]pyrazin-3-yl)urea | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | In Pursuit of an Allosteric Human Tropomyosin Kinase A (hTrkA) Inhibitor for Chronic Pain
Acs Med.Chem.Lett., 2021
|
|
9B7O
 
 | |
9B7P
 
 | |
6JKD
 
 | | Crystal structure of tetrameric PepTSo2 in I4 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
9B7K
 
 | |
1JYN
 
 | |
5VVI
 
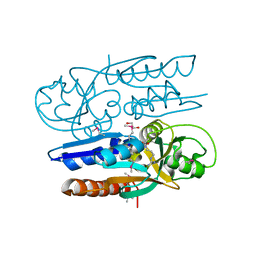 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
6D24
 
 | | Trypanosoma cruzi Glucose-6-P Dehydrogenase in complex with G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Botti, H, Ortiz, C, Comini, M.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Glucose-6-Phosphate Dehydrogenase from the Human Pathogen Trypanosoma cruzi Evolved Unique Structural Features to Support Efficient Product Formation.
J.Mol.Biol., 431, 2019
|
|
6D3I
 
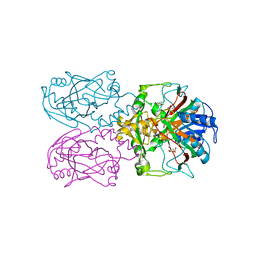 | | ftv7 dioxygenase with 2,4-D bound | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6JER
 
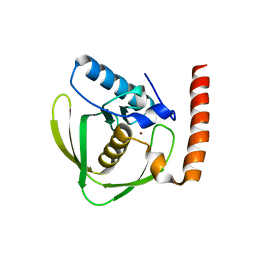 | | Apo crystal structure of class I type a peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, crystallization, and preliminary X-ray crystallographic analysis of peptide deformylase from Acinetobacter baumanii
Biodesign, 5, 2017
|
|
4X65
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6CT6
 
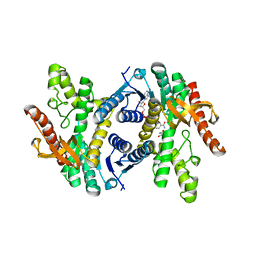 | | Crystal structure of lactate dehydrogenase from Eimeria maxima with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Lactate dehydrogenase, OXAMIC ACID, ... | | Authors: | Wirth, J.D, Xu, C, Theobald, D.L. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | The Mechanistic, Structural, and Evolutionary Origin of Lactate Dehydrogenase Substrate Specificity in Apicomplexa
To Be Published
|
|
8RDN
 
 | | Holomycin methyltransferase DtpM with SAH and XRD-271 | | Descriptor: | CHLORIDE ION, DtpM, GLYCEROL, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isofunctional but Structurally Different Methyltransferases for Dithiolopyrrolone Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7PTQ
 
 | | RNA origami 5-helix tile | | Descriptor: | Chains: C | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
1JZ8
 
 | |
1JZ5
 
 | |
5O3Y
 
 | | SOD1 bound to Ebsulfur | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Capper, M.J, Wright, G.S.A, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation.
Nat Commun, 9, 2018
|
|
7PTS
 
 | | RNA origami 5-helix tile | | Descriptor: | 5HT-B | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.71 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
