5ZZ4
 
 | | Crystal structure of bruton's tyrosine kinase in complex with inhibitor 2e | | Descriptor: | N-[3-(4-amino-6-{[4-(morpholine-4-carbonyl)phenyl]amino}-1,3,5-triazin-2-yl)-2-methylphenyl]-4-tert-butylbenzamide, Tyrosine-protein kinase BTK | | Authors: | Kawahata, W, Asami, T, Irie, T, Kiyoi, T, Taniguchi, H, Asamitsu, Y, Inoue, T, Miyake, T, Sawa, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and Synthesis of Novel Amino-triazine Analogues as Selective Bruton's Tyrosine Kinase Inhibitors for Treatment of Rheumatoid Arthritis.
J. Med. Chem., 61, 2018
|
|
2V9K
 
 | | Crystal structure of human PUS10, a novel pseudouridine synthase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | McCleverty, C.J, Hornsby, M, Spraggon, G, Kreusch, A. | | Deposit date: | 2007-08-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Pus10, a Novel Pseudouridine Synthase.
J.Mol.Biol., 373, 2007
|
|
2V3Z
 
 | |
2VDA
 
 | | Solution structure of the SecA-signal peptide complex | | Descriptor: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | Authors: | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
6AAX
 
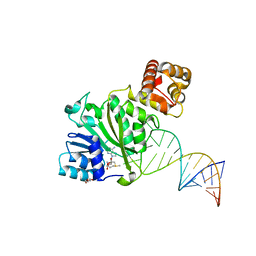 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6ABV
 
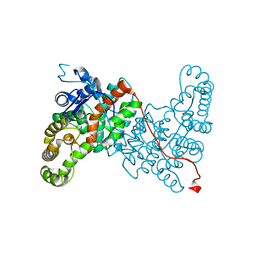 | |
2V6I
 
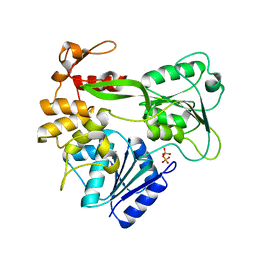 | | Kokobera Virus Helicase | | Descriptor: | PYROPHOSPHATE 2-, RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2VFD
 
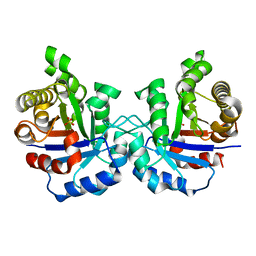 | | Crystal structure of the F96S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6A1K
 
 | | Phosphate acyltransferase PlsX from B.subtilis | | Descriptor: | Phosphate acyltransferase, SULFATE ION | | Authors: | Guo, Z, Jiang, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of an amphipathic peptide sensor of the Bacillus subtilisfluid membrane microdomains.
Commun Biol, 2, 2019
|
|
2VL3
 
 | | Oxidized and reduced forms of human peroxiredoxin 5 | | Descriptor: | PEROXIREDOXIN-5 | | Authors: | Smeets, A, Declercq, J.P. | | Deposit date: | 2008-01-08 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The Crystal Structures of Oxidized Forms of Human Peroxiredoxin 5 with an Intramolecular Disulfide Bond Confirm the Proposed Enzymatic Mechanism for Atypical 2-Cys Peroxiredoxins.
Arch.Biochem.Biophys., 477, 2008
|
|
6A2A
 
 | | Crystal structure of a synthase 2 from santalum album | | Descriptor: | Sesquisabinene B synthase 2 | | Authors: | Han, X, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-09 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a synthase 2 from santalum album
to be published
|
|
2VF3
 
 | | Aquifex aeolicus IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Kinase Ispe: Structure-Activity Relationships and Co-Crystal Structure Analysis.
Org.Biomol.Chem., 6, 2008
|
|
6ADW
 
 | | Crystal structure of the Zika virus NS3 helicase (apo form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Serine protease NS3 | | Authors: | Fang, J, Lu, G, Gong, P. | | Deposit date: | 2018-08-02 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of the Zika Virus NS3 Helicase Help Visualize the Reactant Water Replenishment.
ACS Infect Dis, 5, 2019
|
|
6A30
 
 | | Crystal Structure of Munc13-1 MUN Domain and Synaptobrevin-2 Juxtamembrane Linker Region | | Descriptor: | Protein unc-13 homolog A, Synaptobrevin-2 juxtamembrane linker peptide | | Authors: | Wang, S, Li, Y, Gong, J.H, Ye, S, Yang, X.F, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly.
Nat Commun, 10, 2019
|
|
2V7A
 
 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
2V8W
 
 | | Crystallographic and mass spectrometric characterisation of eIF4E with N7-cap derivatives | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Brown, C.J, Mcnae, I, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and Mass Spectrometric Characterisation of Eif4E with N(7)-Alkylated CAP Derivatives.
J.Mol.Biol., 372, 2007
|
|
6A4J
 
 | | Crystal structure of Thioredoxin reductase 2 from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Bose, M, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
2VHR
 
 | |
6AI2
 
 | |
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
2V3N
 
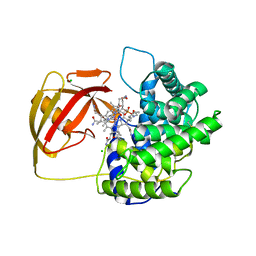 | | Crystallographic analysis of upper axial ligand substitutions in cobalamin bound to transcobalamin | | Descriptor: | CHLORIDE ION, COBALAMIN, CYANIDE ION, ... | | Authors: | Wuerges, J, Geremia, S, Randaccio, L. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Vitamin B12 Transport Proteins: Crystallographic Analysis of Beta-Axial Ligand Substitutions in Cobalamin Bound to Transcobalamin.
Iubmb Life, 59, 2007
|
|
2VCQ
 
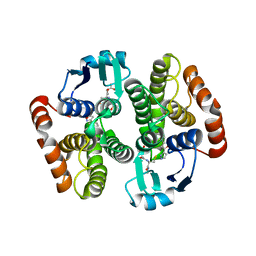 | | Complex structure of prostaglandin D2 synthase at 1.95A. | | Descriptor: | 3-phenyl-5-(1H-pyrazol-3-yl)isoxazole, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2V1R
 
 | | Yeast Pex13 SH3 domain complexed with a peptide from Pex14 at 2.1 A resolution | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20, PEX14 | | Authors: | Kursula, I, Kursula, P, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Genomics of Yeast SH3 Domains
To be Published
|
|
8HKH
 
 | |
2V1U
 
 | | STRUCTURE OF THE AEROPYRUM PERNIX ORC1 PROTEIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*AP*CP*CP*CP*CP*TP*CP*CP*GP*TP *TP*TP*CP*CP*TP*GP*TP*GP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*CP*AP*CP*AP*GP*GP *AP*AP*AP*CP*GP*GP*AP*GP*GP*GP*GP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gaudier, M, Schuwirth, B.S, Westcott, S.L, Wigley, D.B. | | Deposit date: | 2007-05-30 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of DNA Replication Origin Recognition by an Orc Protein.
Science, 317, 2007
|
|
