3K4A
 
 | |
3CE1
 
 | | Crystal Structure of the Cu/Zn Superoxide Dismutase from Cryptococcus liquefaciens Strain N6 | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Teh, A.H, Kanamasa, S, Kajiwara, S, Kumasaka, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Cu/Zn superoxide dismutase from the heavy-metal-tolerant yeast Cryptococcus liquefaciens strain N6.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
6OAG
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
3K4W
 
 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
6OAJ
 
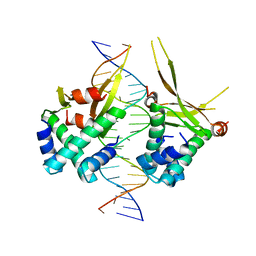 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
6OAM
 
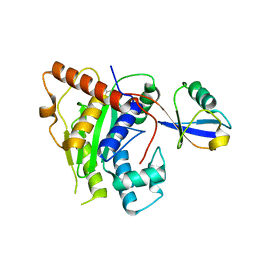 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2019-03-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
6OAQ
 
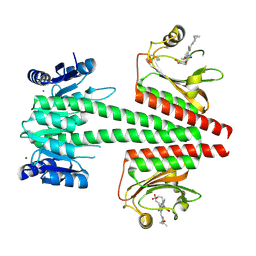 | | Crystal structure of a dual sensor histidine kinase in BeF3- bound state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Dual sensor histidine kinase, MAGNESIUM ION, ... | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OAT
 
 | | Structure of the Ganjam virus OTU bound to sheep ISG15 | | Descriptor: | Interferon stimulated gene 17, RNA-dependent RNA polymerase, prop-2-en-1-amine | | Authors: | Dzimianski, J.V, Williams, I.L, Pegan, S.D. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Determining the molecular drivers of species-specific interferon-stimulated gene product 15 interactions with nairovirus ovarian tumor domain proteases.
Plos One, 14, 2019
|
|
6OB8
 
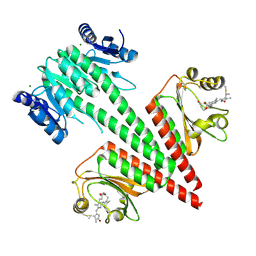 | | Crystal structure of a dual sensor histidine kinase in green light illuminated state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBS
 
 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OC3
 
 | |
3CF8
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with quercetin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3K8M
 
 | | Crystal structure of SusG with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Koropatkin, N.M, Smith, T.J. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SusG: A Unique Cell-Membrane-Associated alpha-Amylase from a Prominent Human Gut Symbiont Targets Complex Starch Molecules.
Structure, 18, 2010
|
|
6OCK
 
 | | Crystal Structure of Leporine Serum Albumin in Complex with Ketoprofen | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, ... | | Authors: | Zielinski, K, Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
6OD1
 
 | | IraD-bound to RssB D58P variant | | Descriptor: | Anti-adapter protein IraD, Regulator of RpoS | | Authors: | Deaconescu, A.M, Dorich, V. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
6OD7
 
 | | Herpes simplex virus type 1 (HSV-1) pUL6 portal protein, dodecameric complex | | Descriptor: | Portal protein | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
6OEP
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3K9I
 
 | |
6OF4
 
 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OFF
 
 | |
3CH7
 
 | |
3K9W
 
 | |
6OFN
 
 | | Crystal structure of green fluorescent protein (GFP); S65T, T203(3-OMeY); ih circular permutant (50-51) | | Descriptor: | Green fluorescent protein (GFP); S65T, T203(3-OMeY); ih circular permutant (50-51) | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3KA3
 
 | | Frog M-ferritin with magnesium | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-18 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
6OGK
 
 | | MeCP2 MBD in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
