2YFH
 
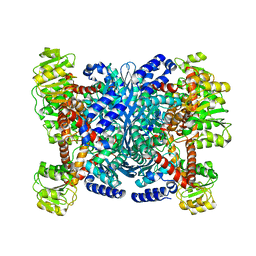 | | Structure of a Chimeric Glutamate Dehydrogenase | | Descriptor: | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Oliveira, T, Panjikar, S, Sharkey, M.A, Carrigan, J.B, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of a Chimaeric Bacterial Glutamate Dehydrogenase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3B0U
 
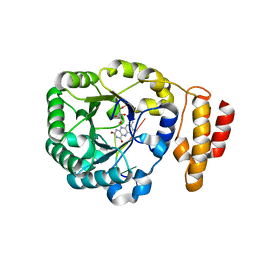 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA fragment | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*(H2U)P*A)-3'), tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B1L
 
 | |
3B1Y
 
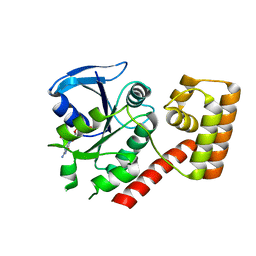 | | Crystal structure of an S. thermophilus NFeoB T35A mutant bound to GDP | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ferrous iron uptake transporter protein B | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3B24
 
 | | Hsp90 alpha N-terminal domain in complex with an aminotriazine fragment molecule | | Descriptor: | 4-(ethylsulfanyl)-6-methyl-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3BI0
 
 | |
3B07
 
 | | Crystal structure of octameric pore form of gamma-hemolysin from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, K, Kawai, Y, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Crystal structure of the octameric pore of
staphylococcal gamma-hemolysin reveals the beta-barrel
pore formation mechanism by two components
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B27
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor Ro4919127 | | Descriptor: | 4-(2-chlorophenyl)-6-(methylsulfanyl)-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3B2K
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B0N
 
 | | Q448K mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B6T
 
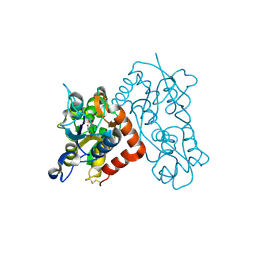 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Cho, Y, Lolis, E, Howe, J.R. | | Deposit date: | 2007-10-29 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
3B8Q
 
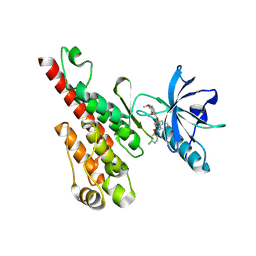 | | Crystal structure of the VEGFR2 kinase domain in complex with a naphthamide inhibitor | | Descriptor: | N-(4-chlorophenyl)-6-[(6,7-dimethoxyquinolin-4-yl)oxy]naphthalene-1-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Gu, Y, Zhao, H. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Naphthamides as Novel and Potent Vascular Endothelial Growth Factor
Receptor Tyrosine Kinase Inhibitors: Design, Synthesis and Evaluation
J.Med.Chem., 51, 2008
|
|
3B25
 
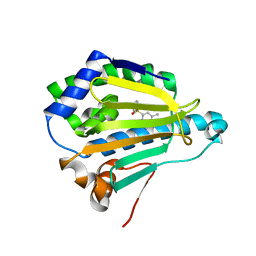 | | Hsp90 alpha N-terminal domain in complex with an inhibitor CH4675194 | | Descriptor: | 4-Methyl-6-(toluene-4-sulfonyl)-pyrimidin-2-ylamine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3B2H
 
 | | Iodide derivative of human LFABP at high resolution | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3BI1
 
 | |
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
3B0G
 
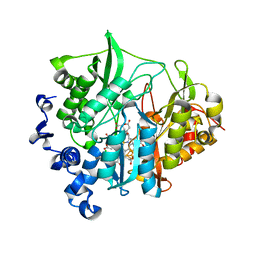 | | Assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B1V
 
 | | Crystal structure of an S. thermophilus NFeoB E67A mutant bound to mGMPPNP | | Descriptor: | 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, CHLORIDE ION, Ferrous iron uptake transporter protein B, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3B2L
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B7D
 
 | |
2ZS0
 
 | | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular giant hemoglobin major globin subunit A1, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Heterotropic and Homotropic Interactions of Invertebrate Giant Hemoglobin
Biochemistry, 47, 2008
|
|
2ZSN
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [300 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
