7S0E
 
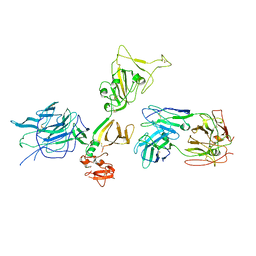 | |
7S0B
 
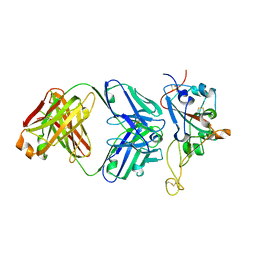 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
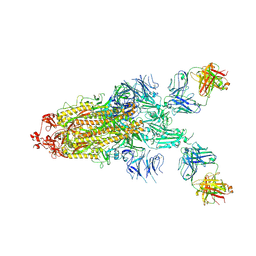
7S0C
 
 | |
7VAH
 
 | |
7RZC
 
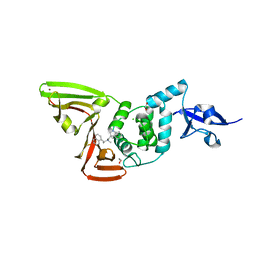 | | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor
To be Published
|
|
7RZU
 
 | |
7RZQ
 
 | |
7RZS
 
 | |
7RZR
 
 | |
7RZT
 
 | |
7RZV
 
 | |
7PKU
 
 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
7RXD
 
 | | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | | Descriptor: | Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V7M
 
 | | crystal structure of SARS-CoV-2 3CL protease | | Descriptor: | 3C-like proteinase | | Authors: | Yi, Y, Zhang, M, Ye, M. | | Deposit date: | 2021-08-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
7RW2
 
 | |
7RVO
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVM
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI11 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVW
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | Descriptor: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVS
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVR
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI18 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI27 | | Descriptor: | 3C-like proteinase, N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
 | |
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVV
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
