6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
7Z3M
 
 | |
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFI
 
 | | SK678 binding region (Siglec + Unique) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
7ZAU
 
 | | Fascin-1 in complex with Nb 3E11 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fascin, ... | | Authors: | Burgess, S.G, Bayliss, R. | | Deposit date: | 2022-03-22 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanobody inhibitor of Fascin-1 actin-bundling activity and filopodia formation.
Open Biology, 14, 2024
|
|
4FUA
 
 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE COMPLEX WITH PGH | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
6LKK
 
 | | Two-component system protein mediate signal transduction | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ABC transporter, solute-binding protein | | Authors: | Wang, M, Tao, Y. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Interface switch mediates signal transmission in a two-component system.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4WFU
 
 | |
6EGK
 
 | | T181N Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
6EER
 
 | | Structure of glycine-bound GoxA from Pseudoalteromonas luteoviolacea | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GoxA, ... | | Authors: | Yukl, E.T, Avalos, D. | | Deposit date: | 2018-08-15 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Spectroscopic Characterization of a Product Schiff Base Intermediate in the Reaction of the Quinoprotein Glycine Oxidase, GoxA.
Biochemistry, 58, 2019
|
|
6EHC
 
 | |
6EHS
 
 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
6EHR
 
 | |
4WF0
 
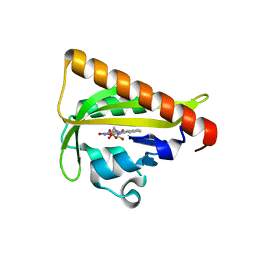 | |
5IEG
 
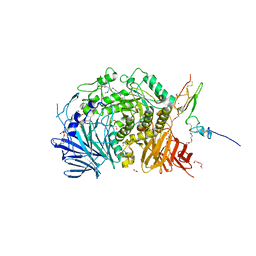 | | Murine endoplasmic reticulum alpha-glucosidase II with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7YPK
 
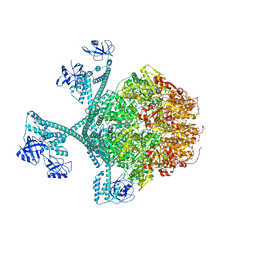 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
6EIA
 
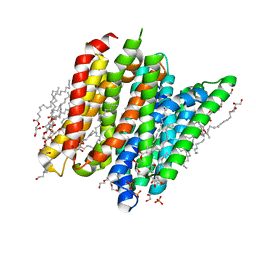 | | PepTSt in complex with HEPES (100 mM) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
4WH1
 
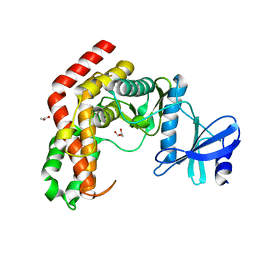 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
6EL1
 
 | | YaxAB pore complex | | Descriptor: | YaxA, YaxB | | Authors: | Braeuning, B, Bertosin, E, Dietz, H, Groll, M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
6EJ1
 
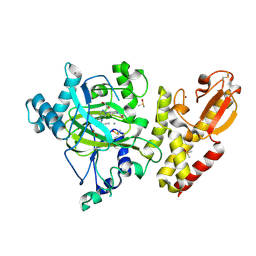 | | Crystal structure of KDM5B in complex with KDOPZ48a. | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-[1-(2-chloranylethanoyl)piperidin-4-yl]pyrazol-4-yl]-7-oxidanylidene-6-propan-2-yl-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ48a.
to be published
|
|
4WIZ
 
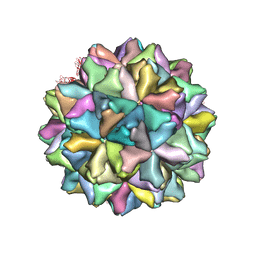 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
6LPP
 
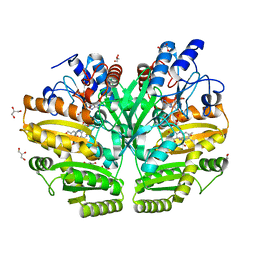 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6EJS
 
 | |
