2PSJ
 
 | |
2PSE
 
 | |
1F4I
 
 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
2PSD
 
 | |
5Y03
 
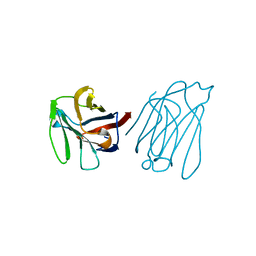 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
7QLL
 
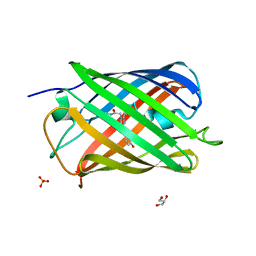 | |
7QLK
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLJ
 
 | |
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
7QLM
 
 | |
7QLN
 
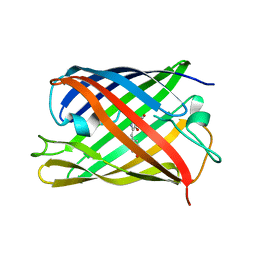 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
3RA7
 
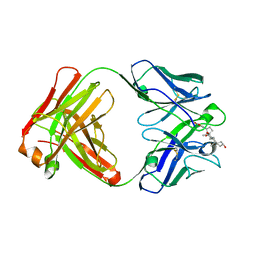 | |
3RWA
 
 | |
3RWT
 
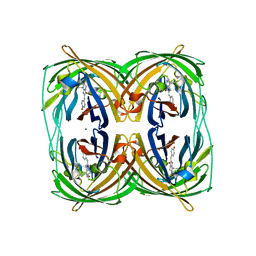 | |
6OAM
 
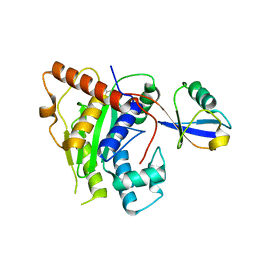 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2019-03-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
2IOV
 
 | | Bright-state structure of the reversibly switchable fluorescent protein Dronpa | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Stiel, A.C, Trowitzsch, S, Weber, G, Andresen, M, Eggeling, C, Hell, S.W, Jakobs, S, Wahl, M.C. | | Deposit date: | 2006-10-11 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants
Biochem.J., 402, 2007
|
|
4EMQ
 
 | | Crystal structure of a single mutant of Dronpa, the green-on-state PDM1-4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Fluorescent protein Dronpa, ... | | Authors: | Ngan, N.B, Van Hecke, K, Van Meervelt, L. | | Deposit date: | 2012-04-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the influence of a single mutation K145N on the oligomerization and photoswitching rate of Dronpa.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6MRN
 
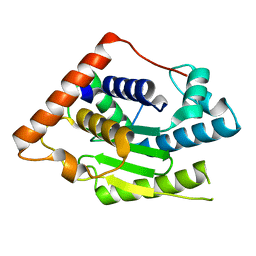 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2 | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
6U95
 
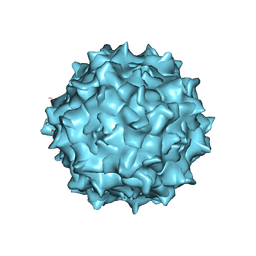 | | Adeno-associated virus strain AAVhu.37 capsid icosahedral structure | | Descriptor: | Capsid protein VP1 | | Authors: | Kaelber, J.T, Yost, S.A, Firlar, E, Mercer, A.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structure of the AAVhu.37 capsid by cryoelectron microscopy.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6U1A
 
 | | Crystal Structure of Fluorescent Protein FusionRed | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Red fluorescent protein | | Authors: | Pletnev, S, Muslinkina, L, Pletneva, N, Pletnev, V.Z. | | Deposit date: | 2019-08-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Two independent routes of post-translational chemistry in fluorescent protein FusionRed.
Int.J.Biol.Macromol., 155, 2020
|
|
5HJN
 
 | |
5HHG
 
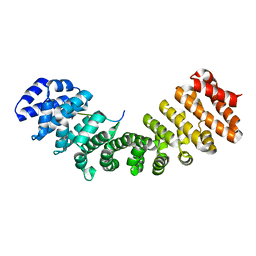 | | Mouse importin alpha: Dengue 2 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, RNA-directed RNA polymerase NS5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
5NOC
 
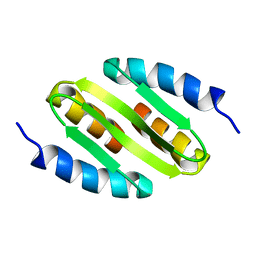 | |
5HJQ
 
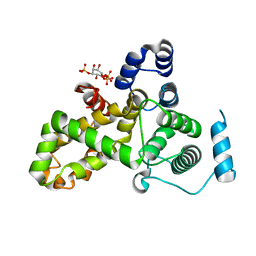 | | Crystal structure of the TBC domain of Skywalker/TBC1D24 from Drosophila melanogaster in complex with inositol(1,4,5)triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, LD10117p | | Authors: | Fischer, B, Paesmans, J, Versees, W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function.
Nat.Struct.Mol.Biol., 23, 2016
|
|
