6KZA
 
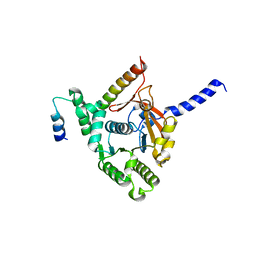 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
9FJP
 
 | |
9FJS
 
 | |
5FZ5
 
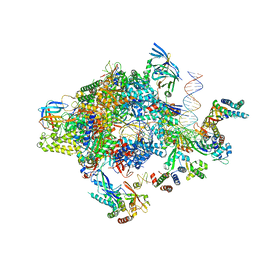 | | Transcription initiation complex structures elucidate DNA opening (CC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
5X6M
 
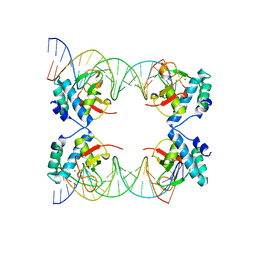 | | Crystal Structure of SMAD5-MH1 in complex with a composite DNA sequence | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
5X6H
 
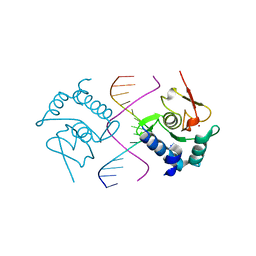 | | Crystal Structure of SMAD5-MH1/GC-BRE DNA complex | | Descriptor: | DNA (5'-D(P*GP*TP*AP*TP*GP*GP*CP*GP*CP*CP*AP*TP*AP*C)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
6FB9
 
 | |
6FB6
 
 | |
6F5B
 
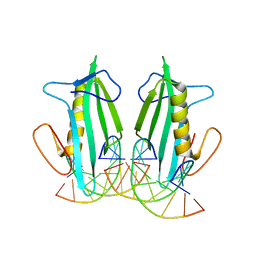 | |
6F5F
 
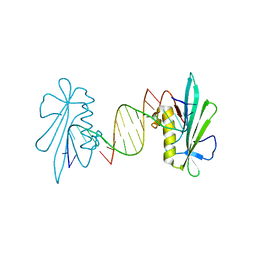 | |
7C17
 
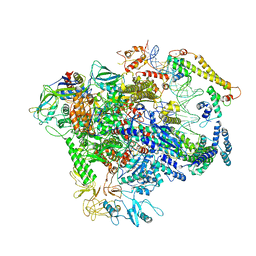 | |
8AV6
 
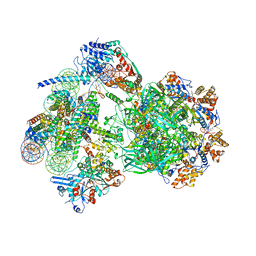 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1TTD
 
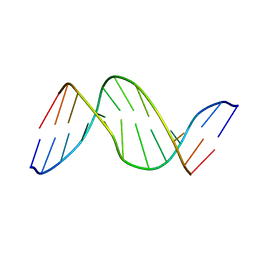 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*(TTD)P*AP*AP*G)-3') | | Authors: | Mcateer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-01-20 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
8ATF
 
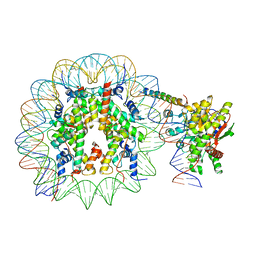 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
5FYW
 
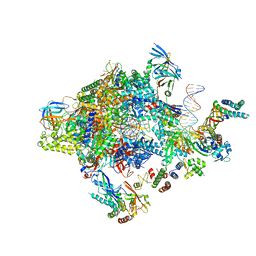 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
6ES2
 
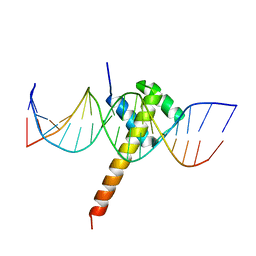 | | Structure of CDX2-DNA(CAA) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
8JH3
 
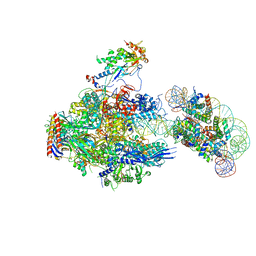 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH4
 
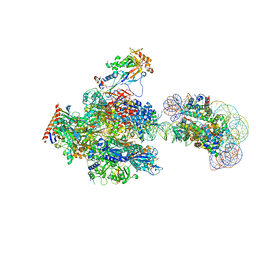 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
1BBX
 
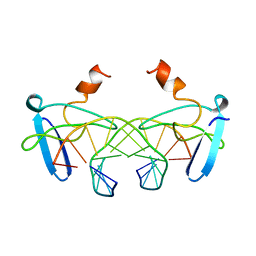 | | NON-SPECIFIC PROTEIN-DNA INTERACTIONS IN THE SSO7D-DNA COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*CP*GP*CP*TP*AP*G)-3'), DNA-BINDING PROTEIN 7D | | Authors: | Agback, P, Baumann, H, Knapp, S, Ladenstein, R, Hard, T. | | Deposit date: | 1998-04-24 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Architecture of Nonspecific Protein-DNA Interactions in the Sso7D-DNA Complex
Nat.Struct.Biol., 5, 1998
|
|
5NSS
 
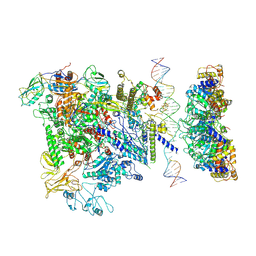 | | Cryo-EM structure of RNA polymerase-sigma54 holoenzyme with promoter DNA and transcription activator PspF intermedate complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
1B3T
 
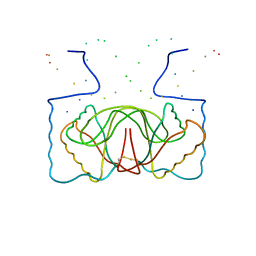 | | EBNA-1 NUCLEAR PROTEIN/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*AP*GP*CP*AP*TP*AP*TP*GP*CP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR PROTEIN EBNA1) | | Authors: | Bochkarev, A, Bochkareva, E, Edwards, A, Frappier, L. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1.
J.Mol.Biol., 284, 1998
|
|
1A1F
 
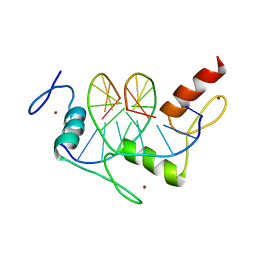 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), THREE-FINGER ZIF268 PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1J
 
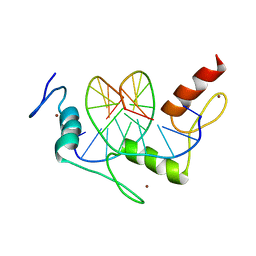 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (RADR ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1H
 
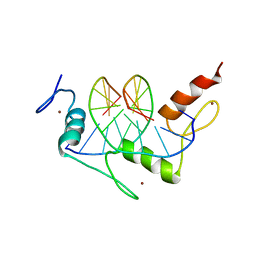 | | QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), QGSR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
