3OKE
 
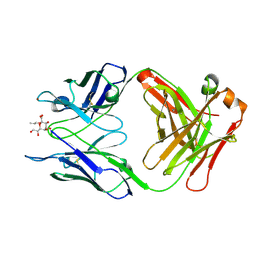 | | Crystal structure of S25-39 in complex with Ko | | Descriptor: | S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ZINC ION, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-24 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
3OMC
 
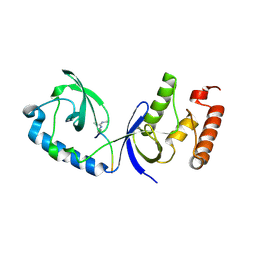 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R4me2s | | Descriptor: | CHLORIDE ION, SYNTHETIC PEPTIDE, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ODA
 
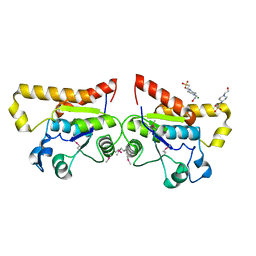 | | Crystal Structure of PSPTO_2114 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | Authors: | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | Deposit date: | 2006-12-22 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
3OKM
 
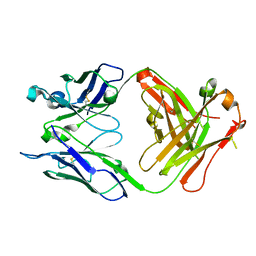 | | Crystal structure of unliganded S25-39 | | Descriptor: | S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2H8K
 
 | | Human Sulfotranferase SULT1C3 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULT1C3 splice variant d | | Authors: | Tempel, W, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and chemical profiling of the human cytosolic sulfotransferases.
Plos Biol., 5, 2007
|
|
3OL4
 
 | |
3OP2
 
 | | Crystal structure of putative mandelate racemase from Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase from
Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate
To be Published
|
|
3OLS
 
 | | Crystal structure of estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Carraz, M, Visser, A, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
3OPM
 
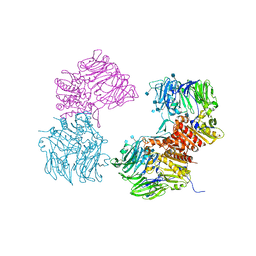 | | Crystal Structure of Human DPP4 Bound to TAK-294 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(aminomethyl)-2-(2-methylpropyl)-1-oxo-4-phenyl-1,2-dihydroisoquinolin-6-yl]oxy}acetamide, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-09-01 | | Release date: | 2011-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of 3-aminomethyl-1,2-dihydro-4-phenyl-1-isoquinolones: a new class of potent, selective, and orally active non-peptide dipeptidyl peptidase IV inhibitors that form a unique interaction with Lys554.
Bioorg.Med.Chem., 19, 2011
|
|
3OJG
 
 | | Structure of an inactive lactonase from Geobacillus kaustophilus with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Tung, A, Robinson, R.C. | | Deposit date: | 2010-08-22 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily
J.Biol.Chem., 285, 2010
|
|
3OTI
 
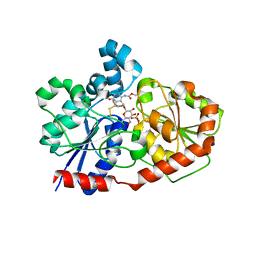 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4G4E
 
 | | Crystal structure of the L88A mutant of HslV from Escherichia coli | | Descriptor: | ATP-dependent protease subunit HslV | | Authors: | Lee, J.W, Park, E, Yoo, H.M, Ha, B.H, An, J.Y, Jeon, Y.J, Seol, J.H, Eom, S.H, Chung, C.H. | | Deposit date: | 2012-07-16 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Structural Alteration in the Pore Motif of the Bacterial 20S Proteasome Homolog HslV Leads to Uncontrolled Protein Degradation
J.Mol.Biol., 425, 2013
|
|
2H70
 
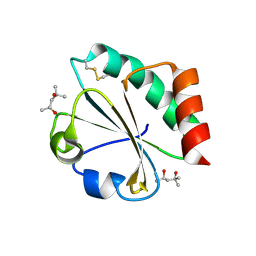 | | Crystal Structure of Thioredoxin Mutant D9E in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
2OBE
 
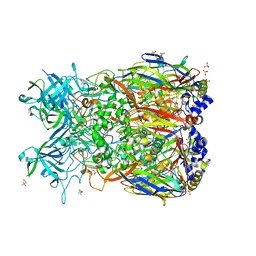 | | Crystal Structure of Chimpanzee Adenovirus (Type 68/Simian 25) Major Coat Protein Hexon | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIHYDROGENPHOSPHATE ION, Hexon protein | | Authors: | Xue, F, Rux, J.J, Burnett, R.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based identification of a major neutralizing site in an adenovirus hexon
J.Virol., 81, 2007
|
|
4G6O
 
 | | Crystal Structure of the ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(aminomethyl)-3-(trifluoromethyl)phenyl]-1H-pyrazol-5-yl}-N-(2,3-dihydro-1-benzofuran-5-ylmethyl)-1H-pyrrole-2-carboxamide, Mitogen-activated protein kinase 1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the ERK2 complexed with E28
to be published
|
|
3OTB
 
 | | Crystal structure of human tRNAHis guanylyltransferase (Thg1) - dGTP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Hyde, S.J, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2010-09-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2HAG
 
 | |
3ON7
 
 | |
2ODR
 
 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4GA0
 
 | |
2HCF
 
 | |
2OEV
 
 | | Crystal structure of ALIX/AIP1 | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2H7G
 
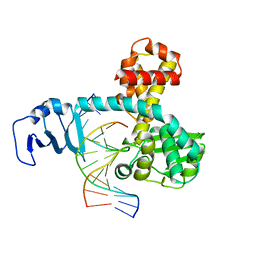 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
3OQ8
 
 | |
