6CK1
 
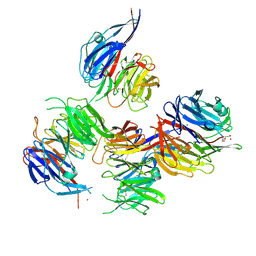 | | Crystal structure of Paracoccus denitrificans AztD | | Descriptor: | A1B2F4 protein, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Yukl, E.T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of AztD provide mechanistic insights into direct zinc transfer between proteins.
Commun Biol, 2, 2019
|
|
5U46
 
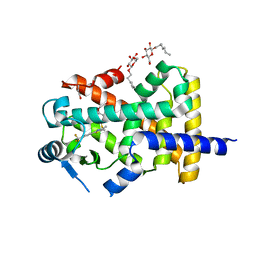 | | Human PPARdelta ligand-binding domain in complexed with GW501516 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, S-1,2-PROPANEDIOL, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BRJ
 
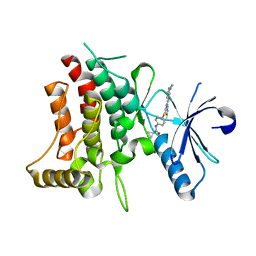 | | DDR1 bound to VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Epithelial discoidin domain-containing receptor 1 | | Authors: | Georghiou, G, Seeliger, M.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | What Makes a Kinase Promiscuous for Inhibitors?
Cell Chem Biol, 26, 2019
|
|
1KX8
 
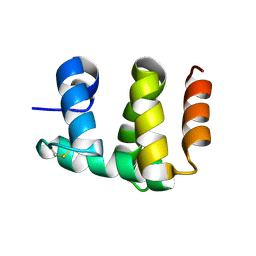 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
5VCB
 
 | |
3QFR
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase (R457H Mutant) | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q5I
 
 | | Crystal Structure of PBANKA_031420 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Panico, E, Crombet, L, Cossar, D, Hassani, A, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of CDPK1 from Plasmodium Bergheii, PBANKA_031420
To be Published
|
|
3QFS
 
 | | Crystal Structure of NADPH-Cytochrome P450 Reductase (FAD/NADPH domain) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH--cytochrome P450 reductase | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5U45
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 14 | | Descriptor: | 6-(2-{[cyclopropyl(3'-fluoro[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1O9O
 
 | |
5U3U
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 5 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-2-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U44
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 13 | | Descriptor: | 6-(2-{[cyclopropyl(2'-fluoro[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, Peroxisome proliferator-activated receptor delta, S-1,2-PROPANEDIOL, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Z5I
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
1Y4H
 
 | | Wild type staphopain-staphostatin complex | | Descriptor: | CHLORIDE ION, SULFATE ION, cysteine protease, ... | | Authors: | Filipek, R, Potempa, J, Bochtler, M. | | Deposit date: | 2004-11-30 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A comparison of staphostatin B with standard mechanism serine protease inhibitors.
J.Biol.Chem., 280, 2005
|
|
5U3Y
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 9 | | Descriptor: | 6-[2-({cyclopropyl[4-(furan-2-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3L30
 
 | | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine | | Descriptor: | 4,14-dihydro-8H-[1,3]dioxolo[4,5-g]isoquino[3,2-a]isoquinoline-9,10-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Naveen, C, Prasanth, G.K, Abhilash, J, Pradeep, M, Ponnuraj, K, Sadasivan, C, Haridas, M. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine
To be Published
|
|
3R8W
 
 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
7MCK
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 18 | | Descriptor: | N-{5-[(3S)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]-2-(trifluoromethyl)pyridin-3-yl}-6-(1-methyl-1H-pyrazol-4-yl)pyridine-2-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Optimization of brain-penetrant picolinamide derived leucine-rich repeat kinase 2 (LRRK2) inhibitors.
Rsc Med Chem, 12, 2021
|
|
5Z5F
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose | | Descriptor: | Beta-xylosidase, CALCIUM ION, beta-L-arabinofuranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
1ZGC
 
 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (RS)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5S)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
7M90
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Y
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M91
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8X
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
