1D3Q
 
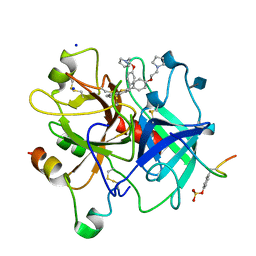 | | CRYSTAL STRUCTURE OF HUMAN ALPHA THROMBIN IN COMPLEX WITH BENZO[B]THIOPHENE INHIBITOR 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-PYRROLIDIN-1-YL-ETHOXY)-BENZYL]-2-4-(2-PYRROLIDIN-1-YL-ETHOXY)-PHENYL] -BENZO[B]THIOPHENE, ALPHA-THROMBIN, ... | | Authors: | Chirgadze, N.Y. | | Deposit date: | 1999-09-30 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of human alpha-thrombin complexed with active site-directed diamino benzo[b]thiophene derivatives: a binding mode for a structurally novel class of inhibitors.
Protein Sci., 9, 2000
|
|
1EMS
 
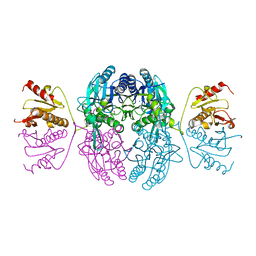 | | CRYSTAL STRUCTURE OF THE C. ELEGANS NITFHIT PROTEIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHYL MERCURY ION, NIT-FRAGILE HISTIDINE TRIAD FUSION PROTEIN, ... | | Authors: | Pace, H.C, Hodawadekar, S.C, Draganescu, A, Huang, J, Bieganowski, P, Pekarsky, Y, Croce, C.M, Brenner, C. | | Deposit date: | 2000-03-17 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
Curr.Biol., 10, 2000
|
|
1EUU
 
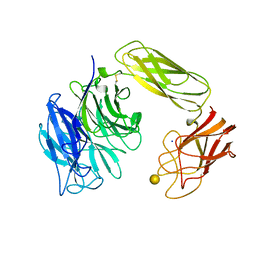 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | Descriptor: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
8C59
 
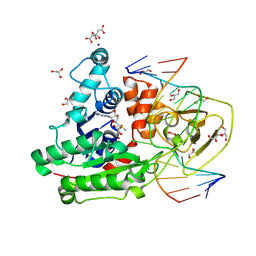 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 2024
|
|
5XAT
 
 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | CITRATE ANION, Citrate-sodium symporter, SODIUM ION, ... | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
8C56
 
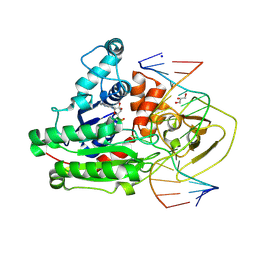 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 2'-deoxy-5-methylzebularine (5mZ) and 5-methylcytosine containing dsDNA | | Descriptor: | Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5PY)P*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 2024
|
|
8CCU
 
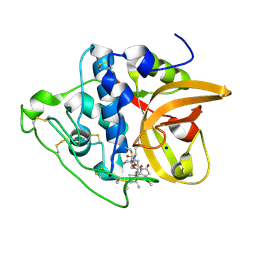 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1
To Be Published
|
|
8CC2
 
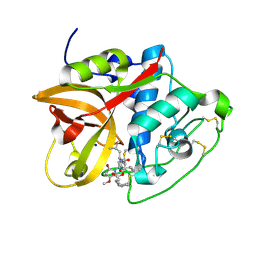 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A
To Be Published
|
|
1SA3
 
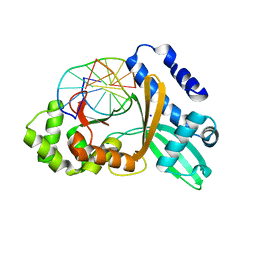 | | An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION, Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.C. | | Deposit date: | 2004-02-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Asymmetric Complex of Restriction Endonuclease MspI on Its Palindromic DNA Recognition Site.
STRUCTURE, 12, 2004
|
|
8CK2
 
 | | MUC2 CysD1 G1352S | | Descriptor: | ACETATE ION, CALCIUM ION, Mucin-2, ... | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pulmonary Fibrosis Mutation Undermines Mucin 5B Supramolecular Assembly
To Be Published
|
|
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
8COD
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Saleem-Batcha, R, Popadic, D, Koeppl, L.H, Andexer, J.N. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine
To Be Published
|
|
8CQV
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
5XAR
 
 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | Citrate-sodium symporter, SODIUM ION, octyl beta-D-glucopyranoside | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
1TW7
 
 | | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target | | Descriptor: | SODIUM ION, protease | | Authors: | Martin, P, Vickrey, J.F, Proteasa, G, Jimenez, Y.L, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2004-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target
Structure, 13, 2005
|
|
5IJS
 
 | | Crystal structure of autotaxin with orthovanadate bound as a trigonal bipyramidal intermediate analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Hausmann, J, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of the catalytic cycle of the phosphodiesterase Autotaxin.
J.Struct.Biol., 195, 2016
|
|
6V1A
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
8CHA
 
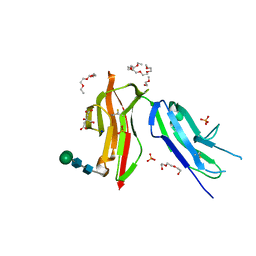 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
2DEK
 
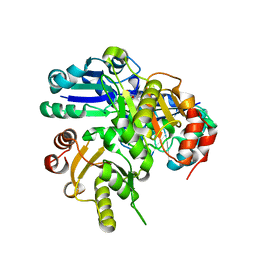 | |
5ISM
 
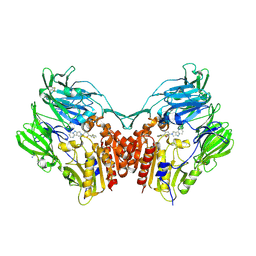 | | Human DPP4 in complex with a novel 5,5,6-tricyclic pyrrolidine inhibitor | | Descriptor: | (2R,3S,5R)-2-(2,5-difluorophenyl)-5-(7H-pyrrolo[3',4':3,4]pyrazolo[1,5-a]pyrimidin-8(9H)-yl)oxan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of novel 5,6,5- and 5,5,6-tricyclic pyrrolidines as potent and selective DPP-4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8DDL
 
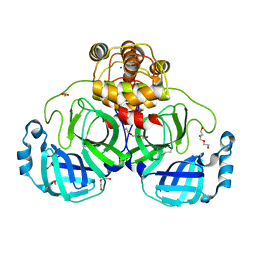 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8CTR
 
 | |
8CW0
 
 | | 20us Temperature-Jump (Light) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWH
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW7
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
