7ZHG
 
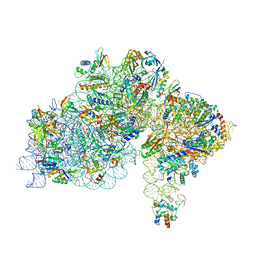 | | High-resolution cryo-EM structure of Pyrococcus abyssi 30S ribosomal subunit bound to mRNA and initiator tRNA anticodon stem-loop | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kazan, R, Bourgeois, G, Mechulam, Y, Coureux, P.D, Schmitt, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7A3M
 
 | |
4NH2
 
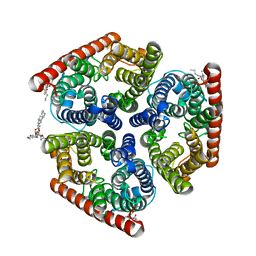 | | Crystal structure of AmtB from E. coli bound to phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Ammonia channel | | Authors: | Laganowsky, A, Reading, E, Allison, T.M, Robinson, C.V. | | Deposit date: | 2013-11-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membrane proteins bind lipids selectively to modulate their structure and function.
Nature, 510, 2014
|
|
1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
2OSX
 
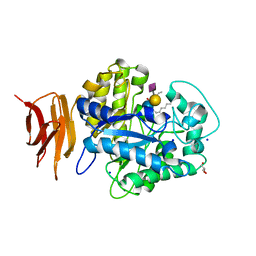 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | Descriptor: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
5FAL
 
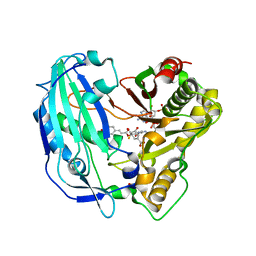 | | Crystal structure of PvHCT in complex with CoA and p-coumaroyl-shikimate | | Descriptor: | (3~{R},4~{R},5~{R})-5-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-3,4-bis(oxidanyl)cyclohexene-1-carboxylic acid, COENZYME A, GLYCEROL, ... | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
9GEU
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione | | Descriptor: | (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione, (R,R)-2,3-BUTANEDIOL, CITRIC ACID, ... | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the human CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,10,11,17,18,21,22-decahydro-4H,16H,20H-13,15-etheno-7,25-(metheno)[1,4]diazepino[2,3-h]pyrido[1,2-p][1]oxa[4,16]diazacyclononadecine-4,8(9H)-dione
To Be Published
|
|
9GEY
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione | | Descriptor: | (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione, 1,2-ETHANEDIOL, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-2-methyl-1,2,3,5,9,10,11,12,18,19,22,23-dodecahydro-17H,21H-14,16-etheno-7,26-(metheno)[1,4]diazepino[2,3-l]pyrido[2,1-d][1]oxa[5,17]diazacycloicosine-4,8-dione
To Be Published
|
|
7AEL
 
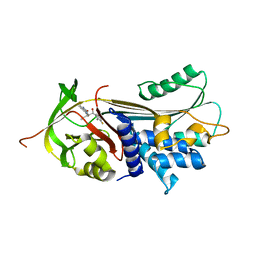 | | alpha 1-antitrypsin (C232S) complexed with GSK716 | | Descriptor: | Alpha-1-antitrypsin, SULFATE ION, ~{N}-[(1~{S},2~{R})-1-(3-fluoranyl-2-methyl-phenyl)-1-oxidanyl-pentan-2-yl]-2-oxidanylidene-1,3-dihydroindole-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of a small molecule that corrects misfolding and increases secretion of Z alpha 1 -antitrypsin.
Embo Mol Med, 13, 2021
|
|
9GET
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)pent-1-en-1-yl)-4-methyl-8-(morpholine-4-carbonyl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one | | Descriptor: | (4~{R})-6-[(~{E})-5-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)pent-1-enyl]-4-methyl-8-morpholin-4-ylcarbonyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Monoglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)pent-1-en-1-yl)-4-methyl-8-(morpholine-4-carbonyl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one
To Be Published
|
|
7SN7
 
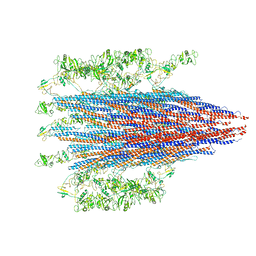 | |
9GGL
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:1 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
9GGN
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:2 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
9GGM
 
 | | Cryo-EM structure of KBTBD4 P313PRR mutant-HDAC2 2:2 complex | | Descriptor: | Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ZINC ION | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
1YWL
 
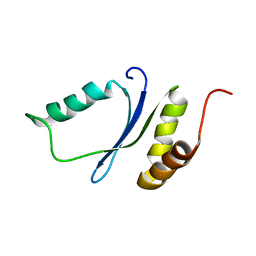 | |
7Z59
 
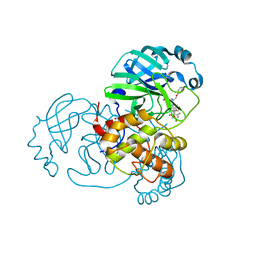 | | SARS-CoV-2 main protease (Mpro) covalently modified with a penicillin derivative | | Descriptor: | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Owen, C.D, Malla, T.R, Brewitz, L, Lukacik, P, Strain-Damerell, C, Mikolajek, H, Muntean, D.G, Aslam, H, Salah, E, Tumber, A, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
J.Med.Chem., 65, 2022
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
7Y1P
 
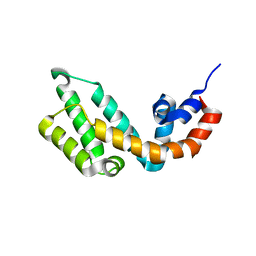 | |
7OVW
 
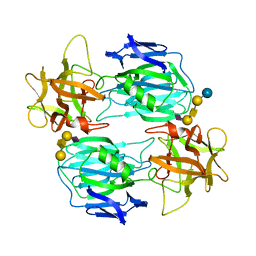 | | Binding domain of botulinum neurotoxin E in complex with GD1a | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type E | | Authors: | Masuyer, G, Stenmark, P. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Ganglioside Receptor Recognition by Botulinum Neurotoxin Serotype E.
Int J Mol Sci, 22, 2021
|
|
4U7U
 
 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
8WB4
 
 | | Structure of PSII-ACPII supercomplex from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
4RZB
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate, SOAKED WITH MERCURY | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-[(E)-iminomethyl]-L-aspartic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An allosteric cyclin E-CDK2 site mapped by paralog hopping with covalent probes.
Nat.Chem.Biol., 21, 2025
|
|
8VQ3
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An allosteric cyclin E-CDK2 site mapped by paralog hopping with covalent probes.
Nat.Chem.Biol., 21, 2025
|
|
