8SJU
 
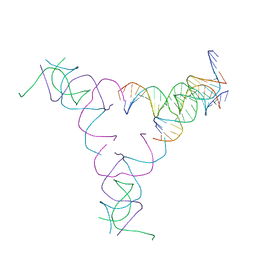 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.14 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJM
 
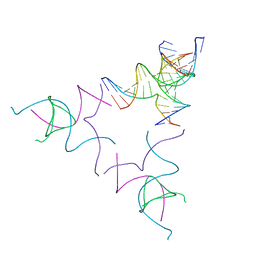 | | [3T12] Self-assembling left-handed tensegrity triangle with 12 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*AP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*TP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
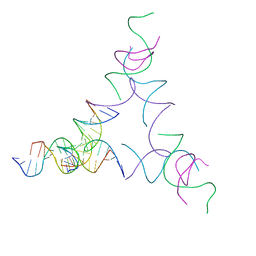 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
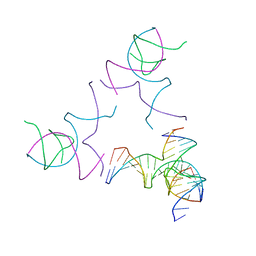 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.3 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJP
 
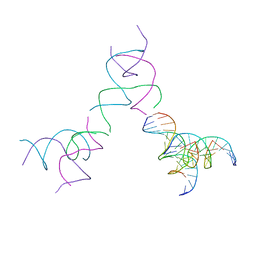 | | [3T15] Self-assembling DNA motif with 15 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*TP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*TP*GP*TP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*TP*GP*GP*AP*CP*AP*GP*GP*GP*G)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.22 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
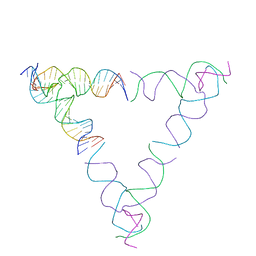 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.64 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJS
 
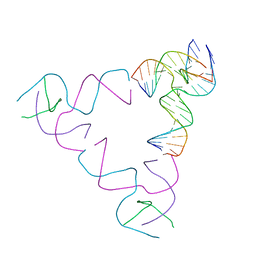 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SL5
 
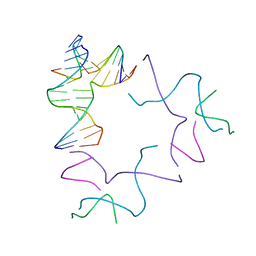 | | [2T13] Self-assembling left-handed two-turn tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.55 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SEI
 
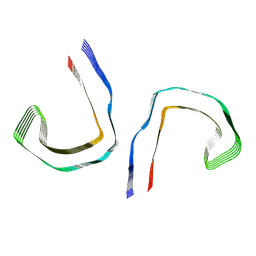 | | SF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S, Bharath, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SJT
 
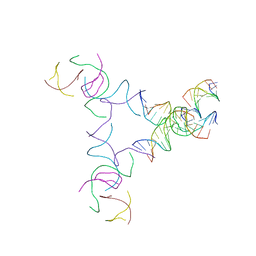 | | [3T14+10] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and a 10 bp linker with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*CP*CP*TP*GP*AP*GP*GP*TP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*AP*CP*TP*CP*TP*GP*CP*TP*A)-3'), DNA (5'-D(*GP*TP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (9.38 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJV
 
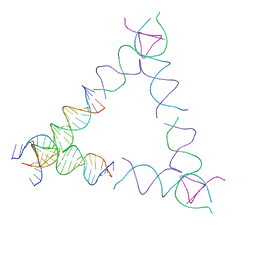 | | [4T24] Self-assembling left-handed tensegrity triangle with 24 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*TP*AP*GP*TP*CP*TP*CP*AP*CP*CP*AP*CP*TP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*GP*AP*AP*CP*AP*CP*TP*CP*CP*TP*GP*AP*GP*AP*CP*TP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*CP*AP*TP*CP*AP*CP*AP*GP*TP*GP*GP*AP*CP*TP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (8.59 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QYG
 
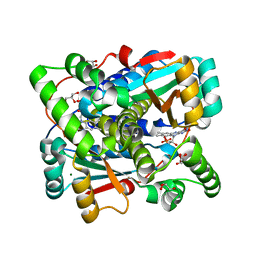 | | Crystal structure of Nitroreductase from Bacillus tequilensis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Russo, S, Fraaije, M.W, Poelarends, G.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Biochemical, kinetic, and structural characterization of a Bacillus tequilensis nitroreductase.
Febs J., 291, 2024
|
|
8QPL
 
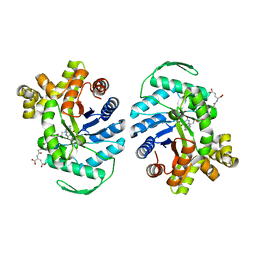 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QFY
 
 | |
1VWA
 
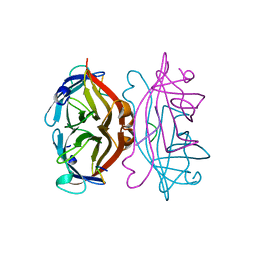 | | STREPTAVIDIN-FSHPQNT | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
8QQ8
 
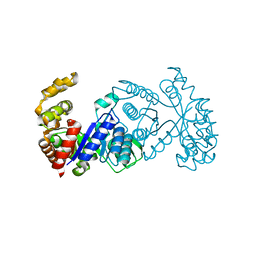 | | Crystal Structure of F420-dependent Methylene-Tetrahydromethanopterin Reductase Mutant E6Q from Methanocaldococcus Jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
1O86
 
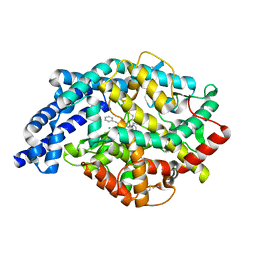 | | Crystal Structure of Human Angiotensin Converting Enzyme in complex with lisinopril. | | Descriptor: | ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, GLYCINE, ... | | Authors: | Natesh, R, Schwager, S.L.U, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2002-11-25 | | Release date: | 2003-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Angiotensin-Converting Enzyme-Lisinopril Complex
Nature, 421, 2003
|
|
8T0E
 
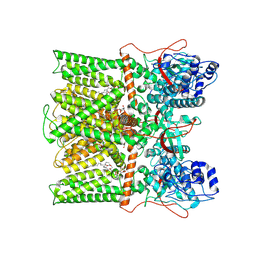 | | TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo) | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Cheng, Y. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5NC9
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2,6-diamino-N-hydroxyhexanamide | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-oxidanyl-hexanamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
8SG2
 
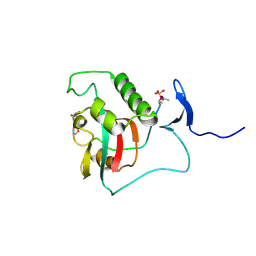 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
8SZ5
 
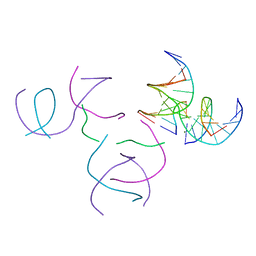 | | [2T5] Self-assembling DNA motif with 5 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5ME9
 
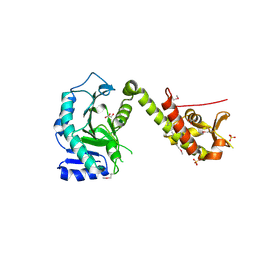 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
8RBX
 
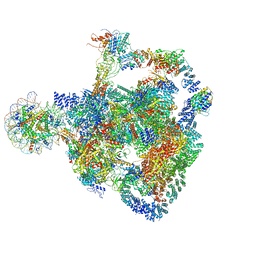 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
1OCV
 
 | |
8ROX
 
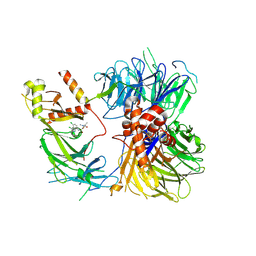 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
