5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JHJ
 
 | | M. Oryzae effector AVR-Pia mutant H3 | | Descriptor: | Antivirulence protein AVR-Pia | | Authors: | Padilla, A, deGuillen, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the Magnaporthe oryzae Effector AVR-Pia by the Decoy Domain of the Rice NLR Immune Receptor RGA5.
Plant Cell, 29, 2017
|
|
5JU7
 
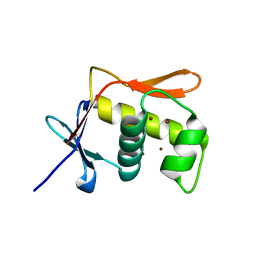 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
4CYK
 
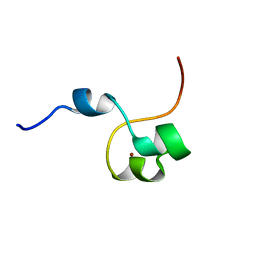 | | Structural basis for binding of Pan3 to Pan2 and its function in mRNA recruitment and deadenylation | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3, ZINC ION | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation.
Embo J., 33, 2014
|
|
7C1M
 
 | | Complex structure of tyrosinated alpha-tubulin carboxy-terminal peptide and A1aY1 binder | | Descriptor: | Carboxy-terminal peptide from tyrosinated alpha-tubulin, Nanobody binder from SSO7d library | | Authors: | Kesarwani, S, Reddy, P.P, Sirajuddin, M, Das, R. | | Deposit date: | 2020-05-05 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Genetically encoded live-cell sensor for tyrosinated microtubules.
J.Cell Biol., 219, 2020
|
|
7BY7
 
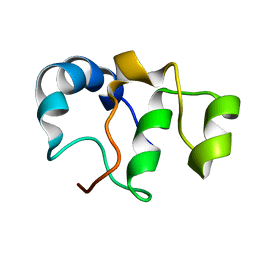 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6NK9
 
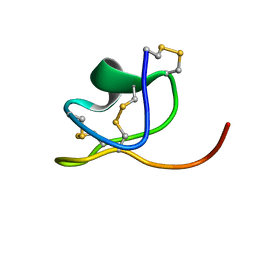 | |
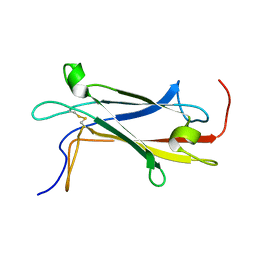
7BCJ
 
 | |
7BUT
 
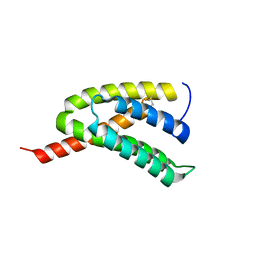 | |
7BW5
 
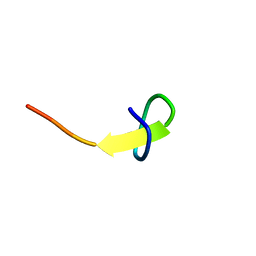 | | a new lasso peptide koreensin | | Descriptor: | lasso peptide koreensin | | Authors: | Hemmi, H, Kodani, S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Heterologous production of new lasso peptide koreensin based on genome mining.
J Antibiot (Tokyo), 74, 2021
|
|
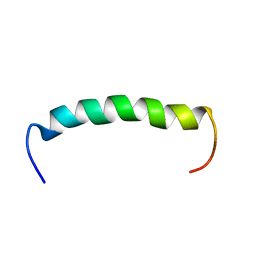
7AT7
 
 | |
7B3J
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
4EQ1
 
 | | Crystal Structure of the ARNT PAS-B homodimer | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Aryl hydrocarbon receptor nuclear translocator | | Authors: | Gardner, K.H, Key, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulating the ARNT/TACC3 Axis: Multiple Approaches to Manipulating Protein/Protein Interactions with Small Molecules.
Acs Chem.Biol., 8, 2013
|
|
5KES
 
 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
5JPW
 
 | | Molecular basis for protein recognition specificity of the DYNLT1/Tctex1 canonical binding groove. Characterization of the interaction with activin receptor IIB | | Descriptor: | Dynein light chain Tctex-type 1,Cytoplasmic dynein 1 intermediate chain 2 | | Authors: | Rodriguez-Crespo, I, Merino-Gracia, J, Bruix, M, Zamora-Carreras, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for the Protein Recognition Specificity of the Dynein Light Chain DYNLT1/Tctex1: CHARACTERIZATION OF THE INTERACTION WITH ACTIVIN RECEPTOR IIB.
J.Biol.Chem., 291, 2016
|
|
1G96
 
 | | HUMAN CYSTATIN C; DIMERIC FORM WITH 3D DOMAIN SWAPPING | | Descriptor: | CHLORIDE ION, CYSTATIN C, GLYCEROL | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Grubb, A, Abrahamson, M, Jaskolski, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping.
Nat.Struct.Biol., 8, 2001
|
|
7B72
 
 | | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(RGT)P*G)-3') | | Authors: | Lomzov, A.A, Pyshnyi, D.V, Shernuykov, A.V, Apukhtina, V.S. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2023-08-23 | | Method: | SOLUTION NMR | | Cite: | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer
To Be Published
|
|
7B71
 
 | | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(SGT)P*G)-3') | | Authors: | Lomzov, A.A, Shernuykov, A.V, Apukhtina, V.S, Pyshnyi, D.V. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer
To Be Published
|
|
7B2B
 
 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2F
 
 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B3K
 
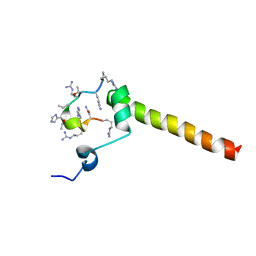 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
1GJS
 
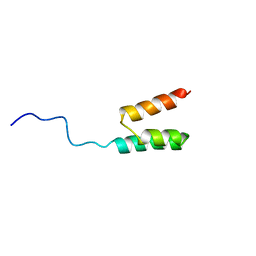 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
6NOX
 
 | | Solution structure of SFTI-KLK5 inhibitor | | Descriptor: | SFTI-KLK5 Peptide | | Authors: | White, A.M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Scanning at P5' within the Bowman-Birk Inhibitory Loop Reveals Specificity Trends for Diverse Serine Proteases.
J. Med. Chem., 62, 2019
|
|
5J2W
 
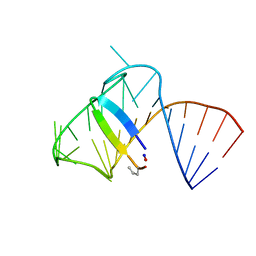 | | Intermediate state lying on the pathway of release of Tat from HIV-1 TAR. | | Descriptor: | Apical region (29mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7ATB
 
 | |
