7KUZ
 
 | |
2R65
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2RD3
 
 | |
2R62
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | Descriptor: | Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
3F42
 
 | | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, protein HP0035 | | Authors: | Chang, C, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori
To be Published
|
|
3FNM
 
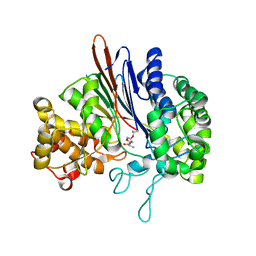 | | Crystal structure of acivicin-inhibited gamma-glutamyltranspeptidase reveals critical roles for its C-terminus in autoprocessing and catalysis | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase (Ggt) Large subunit, Gamma-glutamyltranspeptidase (Ggt) Small subunit | | Authors: | Williams, K, Cullati, S, Sand, A, Biterova, E.I, Barycki, J.J. | | Deposit date: | 2008-12-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Acivicin-Inhibited gamma-Glutamyltranspeptidase Reveals Critical Roles for Its C-Terminus in Autoprocessing and Catalysis.
Biochemistry, 48, 2009
|
|
3GAS
 
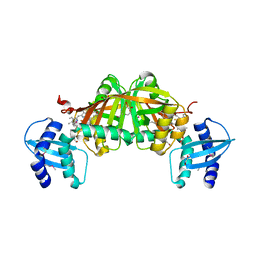 | | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, F, Hu, Y.L, Guo, Y, Guo, G, Zou, Q.M, Wang, D.C. | | Deposit date: | 2009-02-18 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Helicobacter pylori Heme Oxygenase Hugz in Complex with Heme
To be Published
|
|
3IBX
 
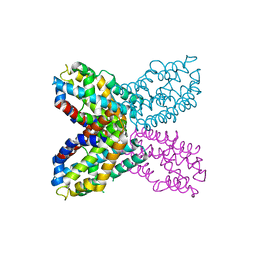 | | Crystal structure of F47Y variant of TenA (HP1287) from Helicobacter pylori | | Descriptor: | Putative thiaminase II | | Authors: | Barison, N, Cendron, L, Trento, A, Angelini, A, Zanotti, G. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity.
Febs J., 276, 2009
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O62
 
 | |
1TK9
 
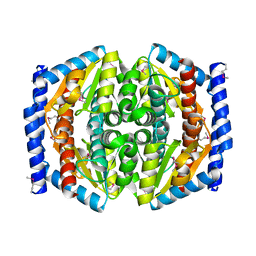 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
3GUQ
 
 | | Crystal structure of novel carcinogenic factor of H. pylori | | Descriptor: | Putative uncharacterized protein | | Authors: | Tsurumura, T, Tsuge, H, Utsunomiya, H, Kise, D, Kuzuhara, T, Fujiki, H, Suganuma, M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for the Helicobacter pylori-carcinogenic TNF-alpha-inducing protein.
Biochem.Biophys.Res.Commun., 388, 2009
|
|
3IEC
 
 | |
3IMP
 
 | |
7T9E
 
 | | Crystal structure of dehaloperoxidase B in complex with +(-)-limonene oxide | | Descriptor: | (1R,4R,6S)-1-methyl-4-(prop-1-en-2-yl)-7-oxabicyclo[4.1.0]heptane, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
7T9C
 
 | | Crystal structure of dehaloperoxidase B in complex with thymol | | Descriptor: | 5-METHYL-2-(1-METHYLETHYL)PHENOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
7T9D
 
 | | Crystal structure of dehaloperoxidase B in complex with -(-) limonene oxide | | Descriptor: | (1S,4S,6R)-1-methyl-4-(prop-1-en-2-yl)-7-oxabicyclo[4.1.0]heptane, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serran, V.S, Ghiladi, R.A, Malewschik, T, Yun, D. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Multifunctional Globin Dehaloperoxidase as a Biocatalyst in the Oxidation of Monoterpenes
To Be Published
|
|
5N6P
 
 | | AMPA receptor NTD mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Rossmann, M, Krieger, J.M. | | Deposit date: | 2017-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric communication pathways in glutamate receptor N-terminal domains
To Be Published
|
|
6GXC
 
 | | Bacterial oligosaccharyltransferase PglB in complex with an inhibitory peptide and a reactive lipid-linked oligosaccharide analog | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLY-ASP-GLN-DAB-ALA-THR-PPN-GLY, ... | | Authors: | Napiorkowska, M, Locher, K.P, Boilevin, J, Darbre, T, Reymond, J.-L. | | Deposit date: | 2018-06-27 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structure of bacterial oligosaccharyltransferase PglB bound to a reactive LLO and an inhibitory peptide.
Sci Rep, 8, 2018
|
|
8GNA
 
 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
5M50
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 3, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-P, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7ADF
 
 | | SFX structure of dehaloperoxidase B in the ferric form | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
7ACP
 
 | | Serial synchrotron structure of dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
6VDR
 
 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Deuteroporphyrin IX and Substrate 4-bromophenol | | Descriptor: | 1,2-ETHANEDIOL, 4-BROMOPHENOL, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
