5X3E
 
 | | kinesin 6 | | Descriptor: | IODIDE ION, Kinesin-like protein, SULFATE ION | | Authors: | Chen, Z, Guan, R, Zhang, L. | | Deposit date: | 2017-02-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of Zen4 in the apo state reveals a missing conformation of kinesin
Nat Commun, 8, 2017
|
|
1QJM
 
 | |
1QNF
 
 | | STRUCTURE OF PHOTOLYASE | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, FLAVIN-ADENINE DINUCLEOTIDE, PHOTOLYASE | | Authors: | Miki, K, Kitadokoro, K. | | Deposit date: | 1997-07-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA photolyase from Anacystis nidulans
Nat.Struct.Biol., 4, 1997
|
|
1QUF
 
 | | X-RAY STRUCTURE OF A COMPLEX NADP+-FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 2.25 ANGSTROMS | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-09-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
5X7U
 
 | | Trehalose synthase from Thermobaculum terrenum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Trehalose synthase | | Authors: | Su, J, Wang, F. | | Deposit date: | 2017-02-27 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum.
J. Agric. Food Chem., 65, 2017
|
|
1QXW
 
 | | Crystal structure of Staphyloccocus aureus in complex with an aminoketone inhibitor 54135. | | Descriptor: | (3S)-3-AMINO-1-(CYCLOPROPYLAMINO)HEPTANE-2,2-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
5X4I
 
 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X86
 
 | | Crystal structure of TMP bound thymidylate kinase from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural and functional roles of dynamically correlated residues in thymidylate kinase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1QY6
 
 | | Structue of V8 Protease from Staphylococcus aureus | | Descriptor: | POTASSIUM ION, serine protease | | Authors: | Prasad, L, Leduc, Y, Hayakawa, K, Delbaere, L.T.J. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1QYD
 
 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | pinoresinol-lariciresinol reductase | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
6I9W
 
 | | Crystal structure of the halohydrin dehalogenase HheG T123G mutant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Putative oxidoreductase | | Authors: | Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Position 123 of halohydrin dehalogenase HheG plays an important role in stability, activity, and enantioselectivity.
Sci Rep, 9, 2019
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
1QMP
 
 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
1QF5
 
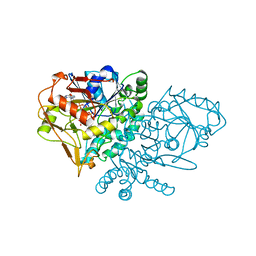 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-S)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Prade, L, Gohda, K, Cowan-Jacob, S.W, Fonne-Pfister, R. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
5XAQ
 
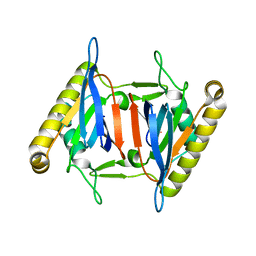 | |
1QA4
 
 | |
5XBQ
 
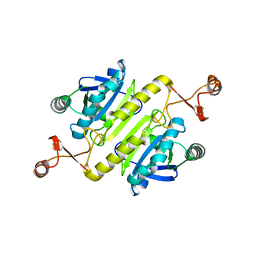 | |
5XB6
 
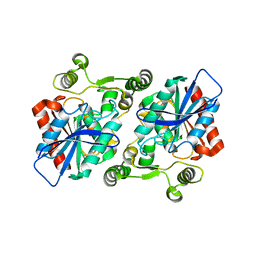 | |
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
6I1Z
 
 | |
5XCP
 
 | |
1QH3
 
 | | HUMAN GLYOXALASE II WITH CACODYLATE AND ACETATE IONS PRESENT IN THE ACTIVE SITE | | Descriptor: | ACETATE ION, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-05-10 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
1QC7
 
 | | T. MARITIMA FLIG C-TERMINAL DOMAIN | | Descriptor: | PROTEIN (FLIG) | | Authors: | Lloyd, S.A, Whitby, F.G, Blair, D, Hill, C.P. | | Deposit date: | 1999-05-18 | | Release date: | 1999-08-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor
Nature, 400, 1999
|
|
5XD5
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with ATP, magnesium fluoride and phosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
5XDM
 
 | |
