1HI7
 
 | |
6I07
 
 | | Crystal structure of EpCAM in complex with scFv | | Descriptor: | Epithelial cell adhesion molecule, GLYCEROL, Single chain Fv | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1HOF
 
 | |
6HGF
 
 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-II: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6HGH
 
 | |
1HNA
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HTB
 
 | |
1HRK
 
 | | CRYSTAL STRUCTURE OF HUMAN FERROCHELATASE | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, FERROCHELATASE | | Authors: | Wu, C.K, Dailey, H.A, Rose, J.P, Burden, A, Sellers, V.M, Wang, B.-C. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
6KV1
 
 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1HNN
 
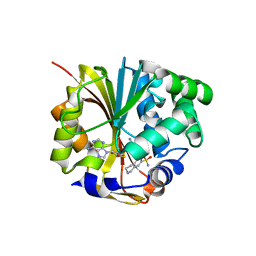 | | CRYSTAL STRUCTURE OF HUMAN PNMT COMPLEXED WITH SK&F 29661 AND ADOHCY(SAH) | | Descriptor: | 1,2,3,4-TETRAHYDRO-ISOQUINOLINE-7-SULFONIC ACID AMIDE, PHENYLETHANOLAMINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Martin, J.L, Begun, J, McLeish, M.J, Caine, J.M, Grunewald, G.L. | | Deposit date: | 2000-12-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT.
Structure, 9, 2001
|
|
1HO9
 
 | |
3Q2F
 
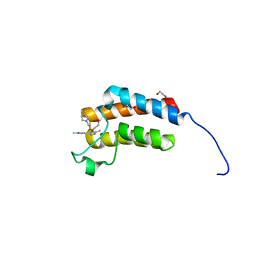 | | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[3-(1H-imidazol-1-yl)propyl]-5-methyl-4H-1,2,4-triazol-3-yl}-1-methyl-1H-pyrazol-5-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Muniz, J.R.C, Filippakopoulos, P, Picaud, S, Felletar, I, Keates, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the bromodomain of human BAZ2B in complex with a triazolo ligand
TO BE PUBLISHED
|
|
1HSQ
 
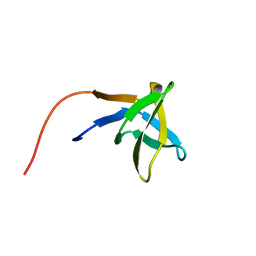 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1HW4
 
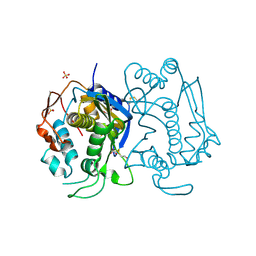 | | STRUCTURE OF THYMIDYLATE SYNTHASE SUGGESTS ADVANTAGES OF CHEMOTHERAPY WITH NONCOMPETITIVE INHIBITORS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Steadman, J.D, Koli, S, Ding, W.C, Minor, W, Dunlap, R.B, Berger, S.H, Lebioda, L. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors.
J.Biol.Chem., 276, 2001
|
|
3BKK
 
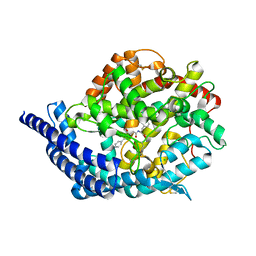 | | Tesis ACE co-crystal structure with ketone ACE inhibitor kAF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Watermeyer, J.M, Kroger, W.L, O'Neill, H.G, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2007-12-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Probing the basis of domain-dependent inhibition using novel ketone inhibitors of Angiotensin-converting enzyme
Biochemistry, 47, 2008
|
|
1HYR
 
 | |
1HOD
 
 | |
5FB6
 
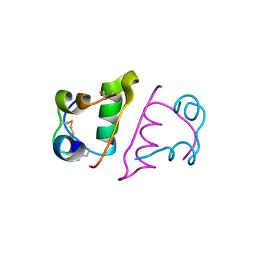 | | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering | | Descriptor: | Insulin Chain A, Insulin Chain B | | Authors: | Roedig, P, Duman, R, Sanchez-Weatherby, J, Vartiainen, I, Burkhardt, A, Warmer, M, David, C, Wagner, A, Meents, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering.
J.Appl.Crystallogr., 49, 2016
|
|
6K1I
 
 | | Human nucleosome core particle with gammaH2A.X variant | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
6JYN
 
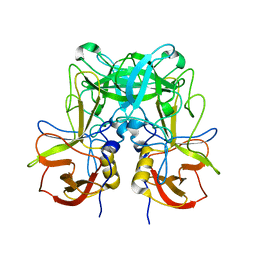 | |
3POO
 
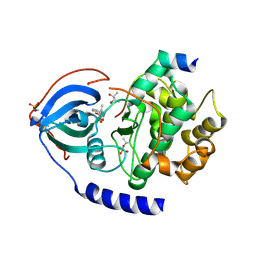 | | human cAMP-dependent protein kinase in complex with an inhibitor | | Descriptor: | (2S,3S)-butane-2,3-diol, N'-[(E)-(2,4-dihydroxy-6-methylphenyl)methylidene]-2-(3-methoxyphenyl)acetohydrazide, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-23 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | fragment based drug design on PKA
To be Published
|
|
6K1K
 
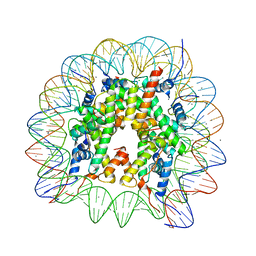 | | Human nucleosome core particle with H2A.X S139E variant | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
3NHE
 
 | |
3AMB
 
 | | Protein kinase A sixfold mutant model of Aurora B with inhibitor VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pflug, A, de Oliveira, T.M, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Biochem.J., 440, 2011
|
|
6I5R
 
 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | Sugar ABC transporter substrate-binding protein, BlMnBP1, ZINC ION, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
