8BWI
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1), crystal form 2 | | Descriptor: | Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
5LFX
 
 | |
5FQE
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
2YGL
 
 | | The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
1XP8
 
 | | Deinococcus radiodurans RecA in complex with ATP-gamma-S | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, RecA protein | | Authors: | Bell, C.E, Rajan, R. | | Deposit date: | 2004-10-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance.
J.Mol.Biol., 344, 2004
|
|
8CBK
 
 | | Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, MAGNESIUM ION, Mitochondrial Precursor tRNA-His(5,Ser), ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
7SIF
 
 | | Crystal Structure of HLA B*3505 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
8R6K
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a phenyltriazine ligand | | Descriptor: | 6-methyl-~{N}-[(5-methylfuran-2-yl)methyl]-3-(4-methylphenyl)-1,2,4-triazin-5-amine, Candida glabrata strain CBS138 chromosome C complete sequence, PHOSPHATE ION, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7SIG
 
 | | Crystal Structure of HLA B*3501 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | Beta-2-microglobulin, CITRATE ANION, MHC class I antigen, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
1X9V
 
 | | Dimeric structure of the C-terminal domain of Vpr | | Descriptor: | VPR protein | | Authors: | Bourbigot, S, Beltz, H, Denis, J, Morellet, N, Roques, B.P, Mely, Y, Bouaziz, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the HIV-1 regulatory protein Vpr adopts an antiparallel dimeric structure in solution via its leucine-zipper-like domain
Biochem.J., 387, 2005
|
|
5NIH
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist LM-12b at 1.3 A resolution. | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
3SRP
 
 | |
3SSX
 
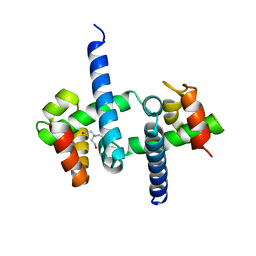 | | E. coli trp aporeporessor L75F mutant | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, Trp operon repressor | | Authors: | Benoff, B, Carey, J, Berman, H.M, Lawson, C.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5801 Å) | | Cite: | Environment-dependent long-range structural distortion in a temperature-sensitive point mutant.
Protein Sci., 21, 2012
|
|
4E2O
 
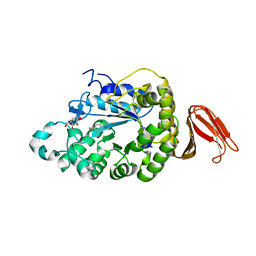 | | Crystal structure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Mok, S.C, Teh, A.H, Saito, J.A, Najimudin, N, Alam, M. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans.
Enzyme.Microb.Technol., 53, 2013
|
|
1E44
 
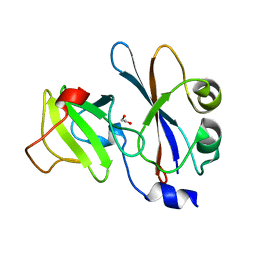 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | Descriptor: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | Authors: | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
1Q8J
 
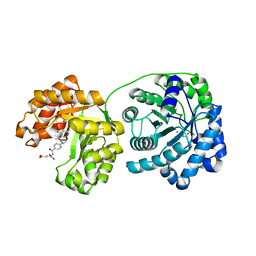 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+, Hcy, methyltetrahydrofolate complex) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, ... | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3T7H
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
8W1Q
 
 | | Aerobic crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
2RRT
 
 | |
7M6O
 
 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
6YVW
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with monocyclic BB-328 | | Descriptor: | 4-[(5-bromanyl-4,6-dimethyl-pyridin-2-yl)amino]-4-oxidanylidene-butanoic acid, Egl nine homolog 1, FE (III) ION, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
3LI2
 
 | | Closed Conformation of HtsA Complexed with Staphyloferrin A | | Descriptor: | (2R)-2-(2-{[(1R)-1-carboxy-4-{[(3S)-3,4-dicarboxy-3-hydroxybutanoyl]amino}butyl]amino}-2-oxoethyl)-2-hydroxybutanedioic acid, ACETATE ION, FE (III) ION, ... | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket.
J.Biol.Chem., 285, 2010
|
|
