6SFA
 
 | |
6SDQ
 
 | |
6T7N
 
 | |
5QPM
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000500a | | Descriptor: | 1-(4-fluorophenyl)-3-(3-hydroxyphenyl)urea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQB
 
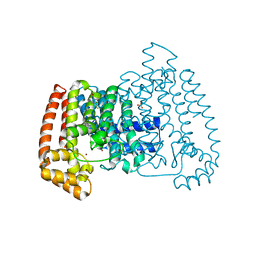 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000676a | | Descriptor: | 8-fluoranyl-5-methyl-1,2,3,6-tetrahydro-1,5-benzodiazocin-4-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPV
 
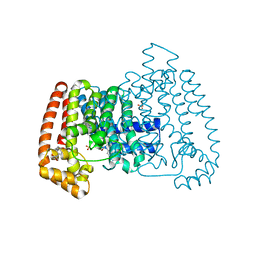 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000416a | | Descriptor: | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPN
 
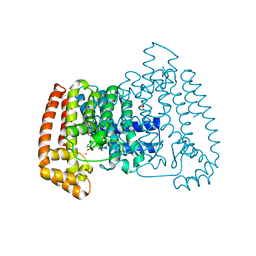 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000576a | | Descriptor: | 3-[(4-methylpiperidin-1-yl)methyl]-1H-indole, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ0
 
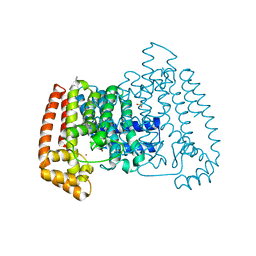 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with XST00000046b | | Descriptor: | 2-morpholin-4-ylaniline, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ6
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000530a | | Descriptor: | (3~{a}~{S},8~{a}~{S})-6-(phenylcarbonyl)-2,3,3~{a},7,8,8~{a}-hexahydropyrrolo[3,4-d]azepin-1-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPX
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000534a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPP
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000512a | | Descriptor: | 1-[2-(trifluoromethyloxy)phenyl]thiourea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ9
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000567a | | Descriptor: | 2-[(2R,5R,6S)-5-hydroxy-1,2,4,5,6,7-hexahydro-3H-2,6-methanoazocino[5,4-b]indol-3-yl]acetamide, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
8CZX
 
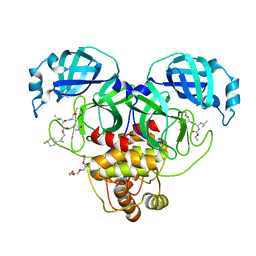 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZW
 
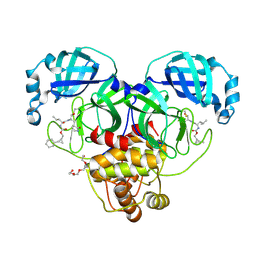 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZV
 
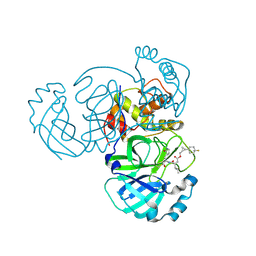 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
5K51
 
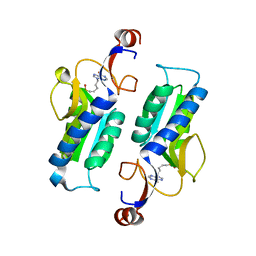 | |
8CBS
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT871 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBR
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8PTL
 
 | | human PHOX2B C-terminal domain including the polyA fragment at 278K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
8PUI
 
 | | human PHOX2B C-terminal domain including the polyA fragment at 298K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
6GEY
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 4g (F125) | | Descriptor: | 2-azanyl-~{N}-[(3,4-dichlorophenyl)methyl]-1,3-benzothiazole-6-carboxamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
7RDB
 
 | | Crystal structure of Tspan15 large extracellular loop (Tspan15 LEL) | | Descriptor: | Tetraspanin-15 | | Authors: | Lipper, C.H, Gabriel, K.H, Seegar, T.C.M, Durr, K.L, Tomlinson, M.G, Blacklow, S.C. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the Tspan15 LEL domain reveals a conserved ADAM10 binding site.
Structure, 30, 2022
|
|
7B4R
 
 | | Structure of the 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus in complex with Coenzyme A and N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Maveyraud, L, Carivenc, C, Blanger, C, Mourey, L. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
