1SZ2
 
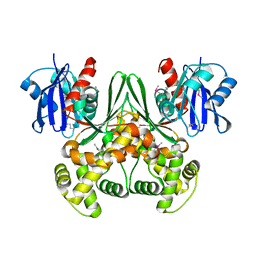 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
7CDH
 
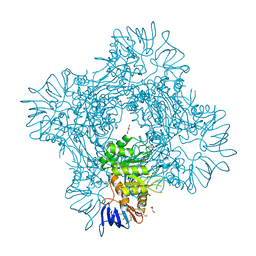 | | Crystal structure of Betaaspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum-AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoaspartyl dipeptidase, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
3NOM
 
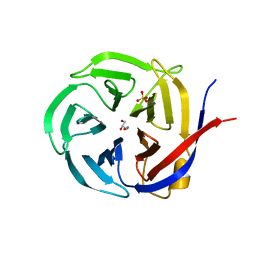 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
7CF6
 
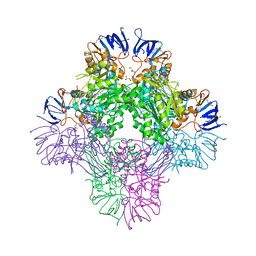 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG4
 
 | | Substrate-free IDE structure in its closed conformation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
2IRU
 
 | |
3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
2IRY
 
 | |
2JG5
 
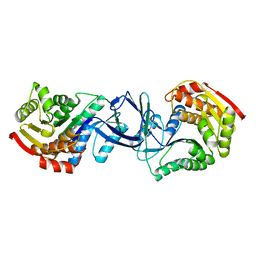 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
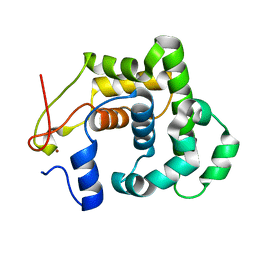 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4B2B
 
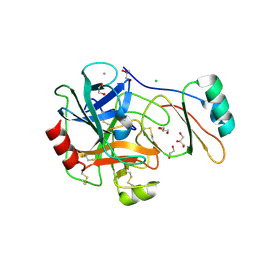 | | Structure of the factor Xa-like trypsin variant triple-Ala (TGPA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
4ABH
 
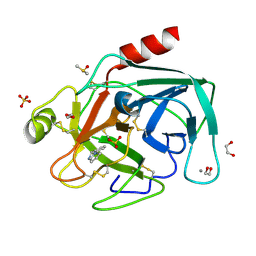 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PYRROLIDIN-1-YLPHENYL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4ABJ
 
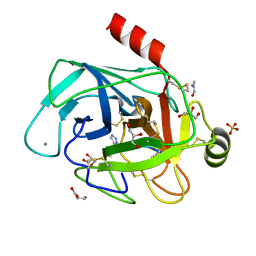 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ABF
 
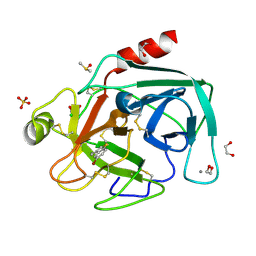 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-BROMO-1-BENZOTHIOPHEN-3-YL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4ABG
 
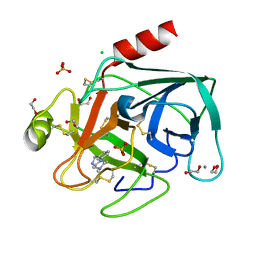 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(4-METHYLPIPERAZIN-1-YL)PHENYL]METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4ABA
 
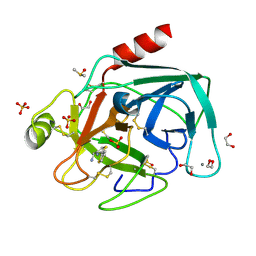 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(thiophen-2-yl)-1,3-thiazol-4-yl]methanamine, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4AB8
 
 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydro-2H-1,5-benzodioxepin-6-ylmethanamine, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4ABE
 
 | |
4AB9
 
 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1-benzofuran-5-yl)methanamine, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4B1T
 
 | | Structure of the factor Xa-like trypsin variant triple-Ala (TA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
4ABB
 
 | |
4ABD
 
 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(1H-pyrrol-1-yl)phenyl]methanamine, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
4B2A
 
 | | Structure of the factor Xa-like trypsin variant triple-Ala (TGA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
9FLA
 
 | |
