7UH3
 
 | | Asp-bound GltPh RSMR mutant in iOFS state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
6GTO
 
 | | Structure of the AtaR antitoxin | | Descriptor: | DUF1778 domain-containing protein, SODIUM ION | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2018-06-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system.
Nat. Chem. Biol., 15, 2019
|
|
8CV3
 
 | |
8CV2
 
 | |
6ZD2
 
 | | Structure of apo telomerase from Candida Tropicalis truncated from C-terminal domain | | Descriptor: | SODIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6CSE
 
 | | Crystal structure of sodium/alanine symporter AgcS with L-alanine bound | | Descriptor: | ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8SZZ
 
 | |
3USJ
 
 | |
2WSY
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
8W6N
 
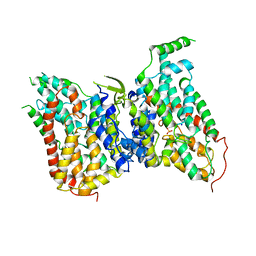 | | NaS1 with sulfate in IN/OUT state | | Descriptor: | SODIUM ION, SULFATE ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
3USO
 
 | |
6H3Z
 
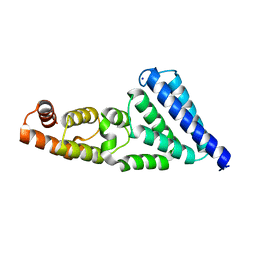 | | Crystal structure of a C-terminal MIF4G domain in NOT1 | | Descriptor: | CCR4-not transcription complex subunit 1, SODIUM ION | | Authors: | Raisch, T, Sandmeir, F, Weichenrieder, O, Valkov, E, Izaurralde, E. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4-NOT complex.
J. Struct. Biol., 204, 2018
|
|
5CD1
 
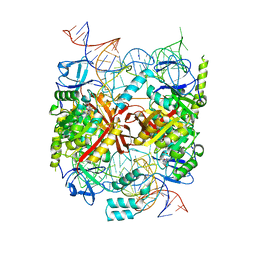 | | Structure of an asymmetric tetramer of human tRNA m1A58 methyltransferase in a complex with SAH and tRNA3Lys | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
6D8G
 
 | |
4ET0
 
 | |
4FY0
 
 | |
2C8L
 
 | | Crystal Structure of (SR) Calcium-ATPase E2(Tg) form | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Jensen, A.M, Sorensen, T.L, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Modulatory and Catalytic Modes of ATP Binding by the Calcium Pump
Embo J., 25, 2006
|
|
6PVN
 
 | |
4F8K
 
 | | Molecular analysis of the interaction between the prostacyclin receptor and the first PDZ domain of PDZK1 | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF3, Prostacyclin receptor | | Authors: | Kocher, O, Birrane, G, Kinsella, B.T, Mulvaney, E.P. | | Deposit date: | 2012-05-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Analysis of the Prostacyclin Receptor's Interaction with the PDZ1 Domain of Its Adaptor Protein PDZK1.
Plos One, 8, 2013
|
|
2M0T
 
 | |
2WG4
 
 | | Crystal structure of the complex between human hedgehog-interacting protein HIP and sonic hedgehog without calcium | | Descriptor: | HEDGEHOG-INTERACTING PROTEIN, SODIUM ION, SONIC HEDGEHOG PROTEIN N-PRODUCT, ... | | Authors: | Bishop, B, Aricescu, A.R, Harlos, K, O'Callaghan, C.A, Jones, E.Y, Siebold, C. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Insights Into Hedgehog Ligand Sequestration by the Human Hedgehog-Interacting Protein Hip
Nat.Struct.Mol.Biol., 16, 2009
|
|
7UGV
 
 | | Asp-bound GltPh RSMR mutant in IFS-A2 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19 F NMR and Cryo-EM.
J.Am.Chem.Soc., 145, 2023
|
|
7UGD
 
 | | Asp-bound GltPh RSMR mutant in IFS-A1 state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, SODIUM ION | | Authors: | Huang, Y, Boudker, O. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Environmentally Ultrasensitive Fluorine Probe to Resolve Protein Conformational Ensembles by 19F NMR and Cryo-EM
J.Am.Chem.Soc., 145, 2023
|
|
5NQK
 
 | | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
2JXO
 
 | | Structure of the second PDZ domain of NHERF-1 | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Cheng, H, Li, J, Dai, Z, Bu, Z, Roder, H. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory Interactions between the PDZ2 and C-terminal Domains in the Scaffolding Protein NHERF1
Structure, 17, 2009
|
|
