3IWV
 
 | | Crystal structure of Y116T mutant of 5-HYDROXYISOURATE HYDROLASE (TRP) | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
5BUL
 
 | | Structure of flavin-dependent brominase Bmp2 triple mutant Y302S F306V A345W | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase triple mutant | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9784 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6M82
 
 | | Crystal structure of TylM1 Y14paF bound to SAH and dTDP-phenol | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.3971 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6MA2
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6AZP
 
 | |
6MCP
 
 | | L. pneumophila effector kinase LegK7 (AMP-PNP bound) in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EOZ
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AsqJ_V72K mutant in complex with cyclopeptin (1b) | | Descriptor: | 2-OXOGLUTARIC ACID, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, NICKEL (II) ION, ... | | Authors: | Groll, M, Braeuer, A, Kaila, V.R.I. | | Deposit date: | 2017-10-10 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism and molecular engineering of quinolone biosynthesis in dioxygenase AsqJ.
Nat Commun, 9, 2018
|
|
5LSZ
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
6MA5
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA4
 
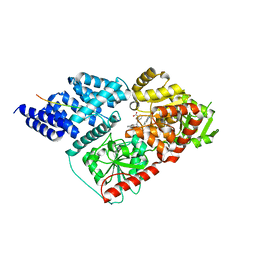 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | Descriptor: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7VT7
 
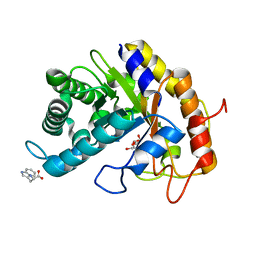 | | Crystal structure of CBM deleted MtGlu5 in complex with CBI | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3IK8
 
 | |
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7VT8
 
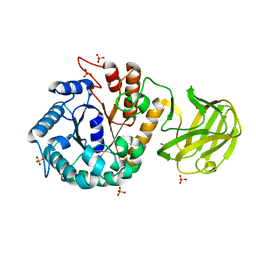 | | Crystal structure of MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, SULFATE ION, beta-D-glucopyranose | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5BS7
 
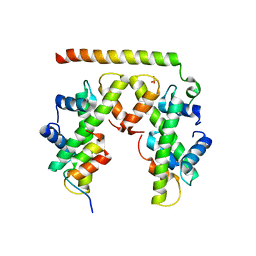 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
6B3S
 
 | |
5LX9
 
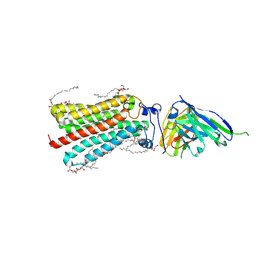 | | CRYSTAL STRUCTURE OF HUMAN ADIPONECTIN RECEPTOR 2 IN COMPLEX WITH A C18 FREE FATTY ACID AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, HUMAN ADIPONECTIN RECEPTOR 2, OLEIC ACID, ... | | Authors: | Vasiliauskaite-Brooks, I, Leyrat, C, Hoh, F, Granier, S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-22 | | Last modified: | 2017-04-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
7VT5
 
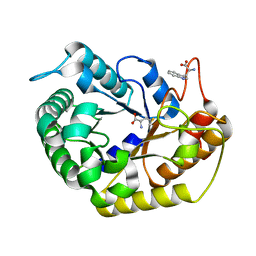 | | Crystal structure of CBM deleted MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, METHIONINE, TRYPTOPHAN | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT6
 
 | | Crystal structure of CBM deleted MtGlu5 in complex with BGC. | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT4
 
 | | Crystal structure of mutant E393Q of MtGlu5 | | Descriptor: | Endoglucanase H, GLYCEROL, SULFATE ION | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6LYN
 
 | | CD146 D4-D5/AA98 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AA98 Fab heavy chain, ... | | Authors: | Chen, X, Yan, X. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146.
Iscience, 24, 2021
|
|
6BE0
 
 | | AvrA delL154 with IP6, CoA | | Descriptor: | AvrA, COENZYME A, INOSITOL HEXAKISPHOSPHATE | | Authors: | Labriola, J.M, Nagar, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural Analysis of the Bacterial Effector AvrA Identifies a Critical Helix Involved in Substrate Recognition.
Biochemistry, 57, 2018
|
|
5M57
 
 | | Nek2 bound to arylaminopurine 6 | | Descriptor: | O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
6LWF
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWC
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
