8OF0
 
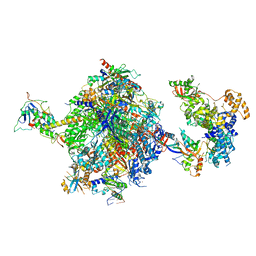 | | Structure of the mammalian Pol II-SPT6-Elongin complex, Structure 1 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEV
 
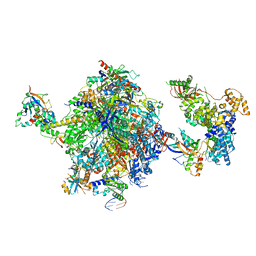 | | Structure of the mammalian Pol II-SPT6-Elongin complex, lacking ELOA latch (composite structure, structure 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEW
 
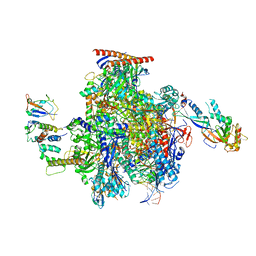 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8Z52
 
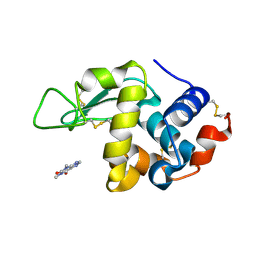 | | Hen Egg-White Lysozyme (HEWL) complexed with Theophylline | | Descriptor: | Lysozyme C, THEOPHYLLINE | | Authors: | Roy, A, Khanppnavar, B, Dolui, S, Datta, S, Maiti, N.C. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hen Egg-White Lysozyme (HEWL) complexed with Theophylline
To Be Published
|
|
8Z51
 
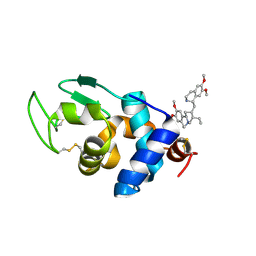 | | Hen Egg-White Lysozyme (HEWL) complexed with Emetine | | Descriptor: | Lysozyme C, emetine | | Authors: | Roy, A, Khanppnavar, B, Dolui, S, Datta, S, Maiti, N.C. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hen Egg-White Lysozyme (HEWL) complexed with Emetine
To Be Published
|
|
8T85
 
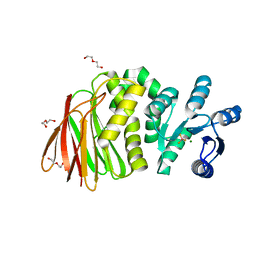 | | Structure of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schwartz, J, Deaconescu, A.M. | | Deposit date: | 2023-06-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of phosphorylated-like RssB, the adaptor delivering sigma s to the ClpXP proteolytic machinery, reveals an interface switch for activation.
J.Biol.Chem., 299, 2023
|
|
8AV8
 
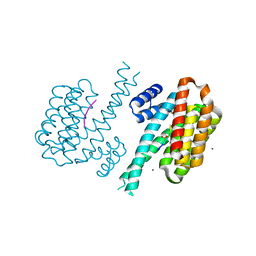 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075300) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenoxycyclopentyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
6KWF
 
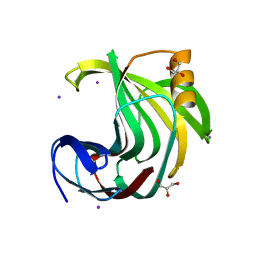 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
4RFC
 
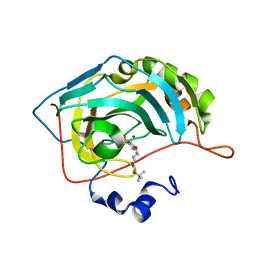 | | Human carbonic anhydrase II in complex with tert-butyl 4-(4-sulfamoylphenoxy)butylcarbamate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, tert-butyl 4-(4-sulfamoylphenoxy)butylcarbamate | | Authors: | Bozdag, M, Pinard, M.A, Carta, F, Masini, E, Scozzafava, A, Mckenna, R, Supuran, C.T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | A class of 4-sulfamoylphenyl-omega-aminoalkyl ethers with effective carbonic anhydrase inhibitory action and antiglaucoma effects.
J.Med.Chem., 57, 2014
|
|
6WRA
 
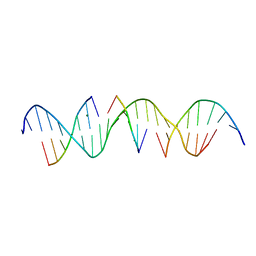 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J34 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6Z7O
 
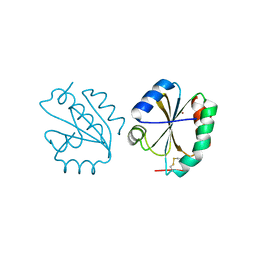 | | Crystal structure of Thioredoxin T from Drosophila melanogaster | | Descriptor: | Thioredoxin-T, ZINC ION | | Authors: | Freier, R, Aragon, E, Baginski, B, Pluta, R, Martin-Malpartida, P, Torner, C, Gonzaez, C, Macias, M. | | Deposit date: | 2020-05-31 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
8A0D
 
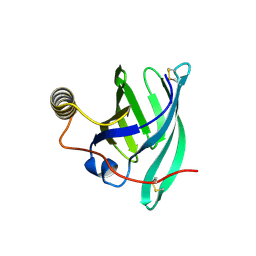 | | Crystal structure of the major guinea pig allergen Cav p 1.0101 part of the lipocalin family | | Descriptor: | Allergen lipocalin Cav p 1 isoform 1 | | Authors: | Herman, R, Charlier, P, Janssen-Weets, B, Hilger, C, Swiontek, K. | | Deposit date: | 2022-05-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.685 Å) | | Cite: | Mammalian derived lipocalin and secretoglobin respiratory allergens strongly bind ligands with potentially immune modulating properties.
Front Allergy, 3, 2022
|
|
6WR9
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J32 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*AP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.071 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRJ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J26 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*A)-3'), DNA (5'-D(P*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSO
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J8 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WR3
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J36 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.173 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRC
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J31 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSQ
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J10 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WST
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J16 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*CP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSN
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J7 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*GP*AP*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WSY
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*AP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-05-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
8RC1
 
 | |
7X5N
 
 | | Crystal structure of E. faecium SHMT in complex with (+)-SHIN-1 and PLP-Ser | | Descriptor: | (4R)-6-azanyl-4-[3-(hydroxymethyl)-5-phenyl-phenyl]-3-methyl-4-propan-2-yl-1H-pyrano[2,3-c]pyrazole-5-carbonitrile, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hasegawa, K, Hayashi, H. | | Deposit date: | 2022-03-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
7R84
 
 | |
4RHF
 
 | | Crystal structure of UbiX mutant V47S from Colwellia psychrerythraea 34H | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, SULFATE ION | | Authors: | Do, H, Kim, S.J, Lee, C.W, Kim, H.-W, Park, H.H, Kim, H.M, Park, H, Park, H.J, Lee, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structure of UbiX, an aromatic acid decarboxylase from the psychrophilic bacterium Colwellia psychrerythraea that undergoes FMN-induced conformational changes.
Sci Rep, 5, 2015
|
|
