3GPQ
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with RNA | | Descriptor: | Non-structural protein 3, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3H3T
 
 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3GTN
 
 | |
3GK8
 
 | | X-ray crystal structure of the Fab from MAb 14, mouse antibody against Canine Parvovirus | | Descriptor: | Fab 14 Heavy Chain, Fab 14 Light Chain | | Authors: | Hafenstein, S, Bowman, V, Sun, T, Nelson, C, Palermo, L, Chipman, P, Battisti, A, Parrish, C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids.
J.Virol., 83, 2009
|
|
3GNZ
 
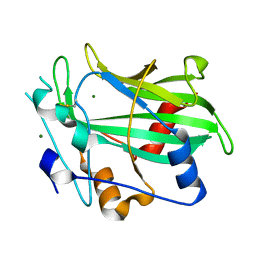 | | Toxin fold for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GZ1
 
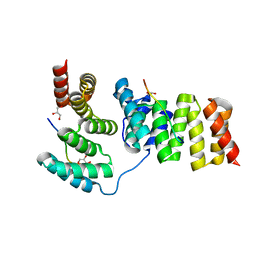 | | Crystal structure of IpgC in complex with the chaperone binding region of IpaB | | Descriptor: | Chaperone protein ipgC, GLYCEROL, Invasin ipaB | | Authors: | Lunelli, M, Lokareddy, R.K, Zychlinsky, A, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | IpaB-IpgC interaction defines binding motif for type III secretion translocator
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GP5
 
 | |
3GVY
 
 | |
3H0D
 
 | |
3GRO
 
 | | Human palmitoyl-protein thioesterase 1 | | Descriptor: | Palmitoyl-protein thioesterase 1, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Seitova, A, Tong, Y, Tempel, W, Dong, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Cossar, D, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Human palmitoyl-protein thioesterase 1
To be Published
|
|
3GSR
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5V peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5V (NLVPVVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3H93
 
 | | Crystal Structure of Pseudomonas aeruginosa DsbA | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein dsbA | | Authors: | Shouldice, S.R. | | Deposit date: | 2009-04-29 | | Release date: | 2009-12-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization of the DsbA Oxidative Folding Catalyst from Pseudomonas aeruginosa Reveals a Highly Oxidizing Protein that Binds Small Molecules.
Antioxid Redox Signal, 12, 2010
|
|
3H9C
 
 | | Structure of methionyl-tRNA synthetase: crystal form 2 | | Descriptor: | CITRIC ACID, Methionyl-tRNA synthetase, ZINC ION | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3GVO
 
 | |
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GKK
 
 | |
3H09
 
 | | The structure of Haemophilus influenzae IgA1 protease | | Descriptor: | ACETATE ION, Immunoglobulin A1 protease, MALONIC ACID, ... | | Authors: | Johnson, T.A, Qiu, J, Plaut, A.G, Holyoak, T. | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Gating Regulates Substrate Selectivity in a Chymotrypsin-Like Serine Protease The Structure of Haemophilus influenzae Immunoglobulin A1 Protease.
J.Mol.Biol., 389, 2009
|
|
3GMY
 
 | |
3GO6
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose and AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE RBSK, ... | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
3GSV
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5Q peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5Q (NLVPQVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3H6W
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | Descriptor: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H81
 
 | |
3GKM
 
 | |
3GLX
 
 | | Crystal Structure Analysis of the DtxR(E175K) complexed with Ni(II) | | Descriptor: | Diphtheria toxin repressor, NICKEL (II) ION, PHOSPHATE ION | | Authors: | D'Aquino, J.A, Denninger, A, Moulin, A, D'Aquino, K.E, Ringe, D. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Decreased sensitivity to changes in the concentration of metal ions as the basis for the hyperactivity of DtxR(E175K).
J.Mol.Biol., 390, 2009
|
|
3H7S
 
 | | Crystal structures of K63-linked di- and tri-ubiquitin reveal a highly extended chain architecture | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Weeks, S.D, Grasty, K.C, Hernandez-Cuebas, L, Loll, P.J. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture.
Proteins, 77, 2009
|
|
