5W62
 
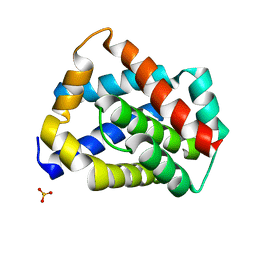 | | Crystal structure of mouse BAX monomer | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
4PKL
 
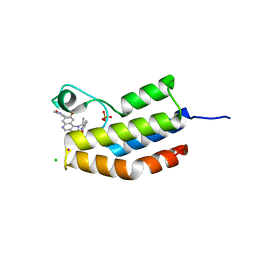 | | Bromodomain of Trypanosoma brucei BDF2 With IBET-151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Bromodomain factor 2, CHLORIDE ION, ... | | Authors: | Debler, E.W. | | Deposit date: | 2014-05-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Bromodomain Proteins Contribute to Maintenance of Bloodstream Form Stage Identity in the African Trypanosome.
Plos Biol., 13, 2015
|
|
6PQN
 
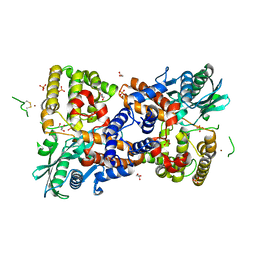 | | Crystal structure of HzTransib transposase | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6SFC
 
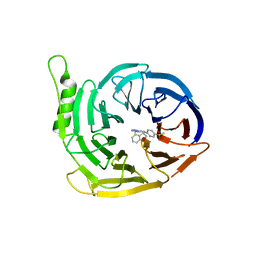 | | EED in complex with a methyl-thiazole | | Descriptor: | CALCIUM ION, N-(1,3-benzodioxol-4-ylmethyl)-4-methyl-5-(1-methylpyrazol-3-yl)-1,3-thiazol-2-amine, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rapid Identification of Novel Allosteric PRC2 Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
4PHD
 
 | |
4X9Z
 
 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
8X9Y
 
 | | E-hexon capsomer of the VZV C-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9W
 
 | | portal vertex capsomer of the VZV C-Capsid | | Descriptor: | CVC1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Nan, W, Lei, C, Jiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
4XIA
 
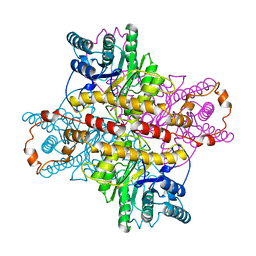 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
1DI0
 
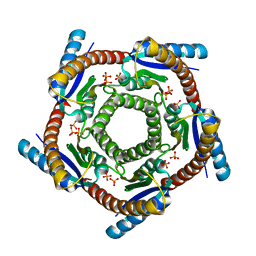 | | CRYSTAL STRUCTURE OF LUMAZINE SYNTHASE FROM BRUCELLA ABORTUS | | Descriptor: | LUMAZINE SYNTHASE, PHOSPHATE ION | | Authors: | Braden, B.C, Velikovsky, C.A, Cauerhff, A.A, Polikarpov, I, Goldbaum, F.A. | | Deposit date: | 1999-11-28 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence in macromolecular assembly: X-ray crystallographic structure analysis of lumazine synthase from Brucella abortus.
J.Mol.Biol., 297, 2000
|
|
3WA5
 
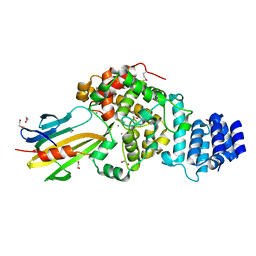 | | Crystal Structure of type VI peptidoglycan muramidase effector Tse3 in complex with its cognate immunity protein Tsi3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Tse3-specific immunity protein, ... | | Authors: | Ding, J, Wang, T, Liu, W, Wang, D.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7V3D
 
 | |
7VMI
 
 | |
6Y3P
 
 | |
6K1D
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
1TXG
 
 | | Structure of glycerol-3-phosphate dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | AMMONIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sakasegawa, S, Hagemeier, C.H, Thauer, R.K, Essen, L.O, Shima, S. | | Deposit date: | 2004-07-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the gpsA gene product of Archaeoglobus fulgidus: A glycerol-3-phosphate dehydrogenase with an unusual NADP+ preference
Protein Sci., 13, 2004
|
|
1U00
 
 | | HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC | | Descriptor: | Chaperone protein hscA, IscU recognition peptide | | Authors: | Cupp-Vickery, J.R, Peterson, J.C, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Molecular Chaperone HscA Substrate Binding Domain Complexed with the IscU Recognition Peptide ELPPVKIHC.
J.Mol.Biol., 342, 2004
|
|
6Y1A
 
 | | Amyloid fibril structure of islet amyloid polypeptide | | Descriptor: | AMINO GROUP, Islet amyloid polypeptide | | Authors: | Roeder, C, Kupreichyk, T, Gremer, L, Schaefer, L.U, Pothula, K.R, Ravelli, R.B.G, Willbold, D, Hoyer, W, Schroder, G.F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-beta fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K18
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1A
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and Mg | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
9GCV
 
 | | Identification of chloride ions in lysozyme at long wavelengths | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | El Omari, K, Forsyth, I, Orr, C.M, Wagner, A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Utilizing anomalous signals for element identification in macromolecular crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6K1C
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
1AK8
 
 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | Descriptor: | CALMODULIN, CERIUM (III) ION | | Authors: | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
8FQO
 
 | | ADC-33 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fish, E.R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
