4NSM
 
 | | crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Collagen-like protein SclB, SULFATE ION | | Authors: | Berisio, R, Squeglia, F, Lukomski, S, Bachert, B. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Streptococcal Collagen-like Protein 2 Globular Domain from Invasive M3-type Group A Streptococcus Shows Significant Similarity to Immunomodulatory HIV Protein gp41.
J.Biol.Chem., 289, 2014
|
|
4O2D
 
 | |
5YKI
 
 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | Descriptor: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
6N93
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2E)-2-(hydroxyimino)propanoic acid, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
4LYQ
 
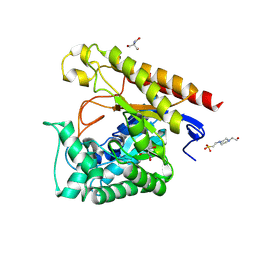 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BAY
 
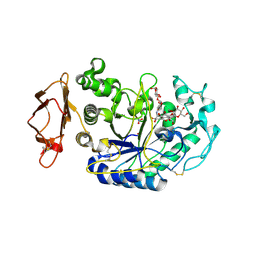 | | N298S Variant of Human Pancreatic Alpha-Amylase in Complex with Nitrate and Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-08 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3BAX
 
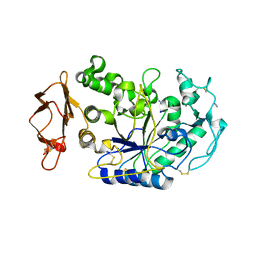 | |
3BAK
 
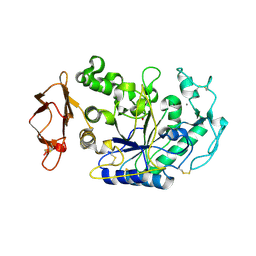 | | N298S mutant of Human Pancreatic Alpha-Amylase in complex with nitrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRATE ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-08 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
4NEY
 
 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR277 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR277, tetrabutylphosphonium | | Authors: | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-30 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR277
To be published
|
|
4NEZ
 
 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR276 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR276, tetrabutylphosphonium | | Authors: | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-30 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR276
To be published
|
|
4NFY
 
 | |
4ELE
 
 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(propan-2-yl)phthalazin-2(1H)-yl]prop-2-en -1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4MDO
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7P7P
 
 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
3BAI
 
 | | Human Pancreatic Alpha Amylase with Bound Nitrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRATE ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
5D8H
 
 | |
4MPM
 
 | | Wild-type human neuroglobin | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Guimaraes, B.G, Golinelli-Pimpaneau, B. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5A8C
 
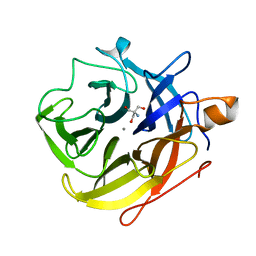 | | The ultra high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 with bound trimethyl N-Oxide (TRS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4N2R
 
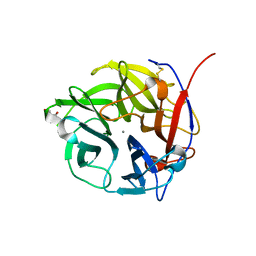 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maydis in complex with L-arabinofuranose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, alpha-L-arabinofuranose, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4MQ4
 
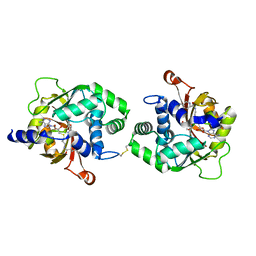 | | Crystal Structure of hPNMT in Complex with bisubstrate inhibitor N-(3-((((2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)thio)propyl)-1,2,3,4-tetrahydroisoquinoline-3-carboxamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-S-(3-{[(3R)-1,2,3,4-tetrahydroisoquinolin-3-ylcarbonyl]amino}propyl)-5'-thioadenosine, Phenylethanolamine N-methyltransferase | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2013-09-15 | | Release date: | 2014-10-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2042 Å) | | Cite: | To be Published
To be Published
|
|
3BAJ
 
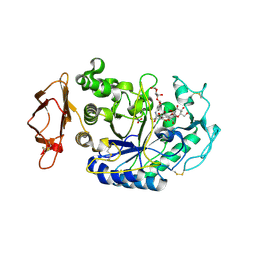 | | Human Pancreatic Alpha-Amylase in Complex with Nitrate and Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
4ODW
 
 | | Unliganded Fab structure of lipid A-specific antibody A6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A6 Fab (IgG2b kappa) light chain, A6 Fab (IgG2b) heavy chain | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2014-01-10 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis for Antibody Recognition of Lipid A: INSIGHTS TO POLYSPECIFICITY TOWARD SINGLE-STRANDED DNA.
J.Biol.Chem., 290, 2015
|
|
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
4LQ0
 
 | |
4MB1
 
 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
