2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2ZWT
 
 | | Crystal Structure of Ferric Cytochrome P450cam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Sakurai, K, Shimada, H, Hayashi, T, Tsukihara, T. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate binding induces structural changes in cytochrome P450cam
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6HEE
 
 | | Crystal structure of Extracellular Domain 1 (ECD1) of FtsX from S. pneumonie in complex with undecyl-maltoside | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsX, SULFATE ION, ... | | Authors: | Martinez-Caballero, S, Alcorlo-Pages, M, Hermoso, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
Mbio, 10, 2019
|
|
4YH5
 
 | | Lipomyces starkeyi levoglucosan kinase bound to ADP and Manganese | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Levoglucosan kinase, ... | | Authors: | Bacik, J.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Producing Glucose 6-Phosphate from Cellulosic Biomass: STRUCTURAL INSIGHTS INTO LEVOGLUCOSAN BIOCONVERSION.
J.Biol.Chem., 290, 2015
|
|
3WCC
 
 | | The complex structure of TcSQS with ligand, E5700 | | Descriptor: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Farnesyltransferase, putative | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
6HPA
 
 | |
6KA0
 
 | | Silver-bound E.coli Malate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
2X6Y
 
 | | Tailspike protein mutant D339A of E.coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAIL SPIKE PROTEIN, ... | | Authors: | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2010-02-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
6K4T
 
 | | Crystal structure of SMB-1 metallo-beta-lactamase in a complex with TSA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-sulfanylbenzoic acid, Metallo-beta-lactamase, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
6HG8
 
 | | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Ambrus, A, Szabo, E, Weichsel, A, Bui, D, Wilk, P, Torocsik, B, Weiss, M.S, Montfort, W.R, Jordan, F, Adam-Vizi, V. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase.
To Be Published
|
|
2X46
 
 | | Crystal Structure of SeMet Arg r 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALLERGEN ARG R 1 | | Authors: | Paesen, G.C, Siebold, C, Syme, N, Harlos, K, Graham, S.C, Hilger, C, Homans, S.W, Hentges, F, Stuart, D.I. | | Deposit date: | 2010-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the Allergen Arg R 1, a Histamine-Binding Lipocalin from a Soft Tick
To be Published
|
|
4YEL
 
 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
2ZFK
 
 | | Crystal Structure of the Kif1A Motor Domain during Mg release: Mg-releasing Transition-2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2008-01-08 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6HCW
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-lysine and a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2ZAL
 
 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
4WHR
 
 | | Anhydride reaction intermediate trapped in Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-fluorobenzene-1,2-diol, 4-fluorooxepine-2,7-dione, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
2NTR
 
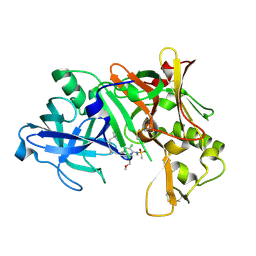 | | Crystal structure of Human Bace-1 bound to inhibitor | | Descriptor: | (2R)-2-(5-{3-chloro-6-((2-methoxyethyl){[(1S,2S)-2-methylcyclopropyl]methyl}amino)-2-[methyl(methylsulfonyl)amino]pyrid in-4-yl}-1,3,4-oxadiazol-2-yl)-1-phenylpropan-2-amine, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-11-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-secretase (BACE-1) inhibitors: accounting for 10s loop flexibility using rigid active sites.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
4WKX
 
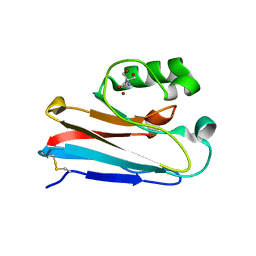 | |
2VHA
 
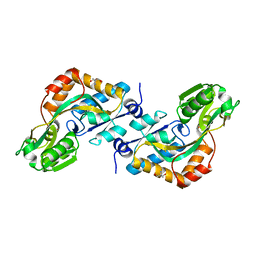 | | DEBP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | Authors: | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | Deposit date: | 2007-11-20 | | Release date: | 2008-07-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
6JHP
 
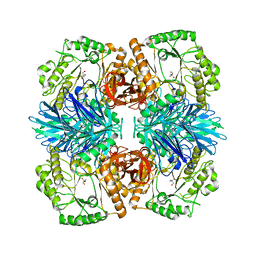 | |
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
7JNY
 
 | | Crystal structure of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Rajasekaran, D, Murphy, J.W, Pantouris, G, Lolis, E.J. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JBS
 
 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
