1LK3
 
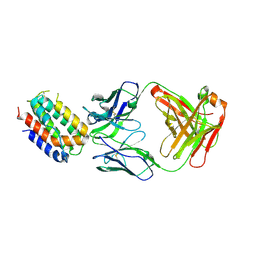 | | ENGINEERED HUMAN INTERLEUKIN-10 MONOMER COMPLEXED TO 9D7 FAB FRAGMENT | | Descriptor: | 9D7 Heavy Chain, 9D7 Light Chain, Interleukin-10 | | Authors: | Josephson, K, Jones, B.C, Walter, L.J, DiGiacomo, R, Indelicato, S.R, Walter, M.R. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Noncompetitive antibody neutralization of IL-10 revealed by protein engineering and x-ray crystallography.
Structure, 10, 2002
|
|
4XEO
 
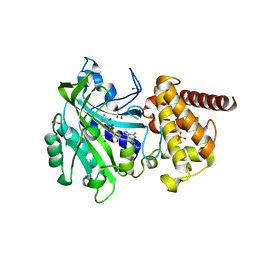 | |
1LLZ
 
 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: reduced enzyme | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, Ferredoxin-dependent glutamate synthase | | Authors: | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
1L76
 
 | |
8EMM
 
 | |
1LN3
 
 | | Structure of Human Phosphatidylcholine Transfer Protein in Complex with Palmitoyl-Linoleoyl Phosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
4XJE
 
 | |
1L8G
 
 | | Crystal structure of PTP1B complexed with 7-(1,1-Dioxo-1H-benzo[d]isothiazol-3-yloxymethyl)-2-(oxalyl-amino)-4,7-dihydro-5H-thieno[2,3-c]pyran-3-carboxylic acid | | Descriptor: | 7-(1,1-DIOXO-1H-BENZO[D]ISOTHIAZOL-3-YLOXYMETHYL)-2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Iversen, L.F, Andersen, H.S, Moller, K.B, Olsen, O.H, Peters, G.H, Branner, S, Mortensen, S.B, Hansen, T.K, Lau, J, Ge, Y, Holsworth, D.D, Newman, M.J, Moller, N.P.H. | | Deposit date: | 2002-03-20 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric hindrance as a basis for structure-based design of selective inhibitors of protein-tyrosine phosphatases.
Biochemistry, 40, 2001
|
|
1L8T
 
 | |
1L91
 
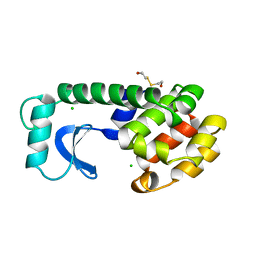 | |
1LO7
 
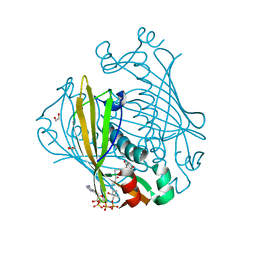 | | X-ray structure of 4-Hydroxybenzoyl CoA Thioesterase complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1L96
 
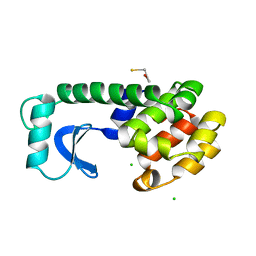 | | STRUCTURE OF A HINGE-BENDING BACTERIOPHAGE T4 LYSOZYME MUTANT, ILE3-> PRO | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Dixon, M, Shewchuk, L, Matthews, B.W. | | Deposit date: | 1992-02-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile3-->Pro.
J.Mol.Biol., 227, 1992
|
|
6UZU
 
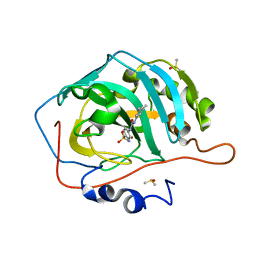 | | Carbonic Anhydrase IX-mimic In Complex WITH U-CH3 | | Descriptor: | 4-{[(3,5-dimethylphenyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | McKenna, R, Mboge, M.Y, Mahon, B.P. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure activity study of carbonic anhydrase IX: Selective inhibition with ureido-substituted benzenesulfonamides.
Eur J Med Chem, 132, 2017
|
|
5IQT
 
 | | WelO5 bound to Fe(II), Cl, 2-oxoglutarate, and 12-epifischerindole U | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Mitchell, A.J, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
1L9E
 
 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1L9W
 
 | | CRYSTAL STRUCTURE OF 3-DEHYDROQUINASE FROM SALMONELLA TYPHI COMPLEXED WITH REACTION PRODUCT | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase aroD | | Authors: | Lee, W.H, Perles, L.A, Nagem, R.A.P, Shrive, A.K, Hawkins, A, Sawyer, L, Polikarpov, I. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6V2N
 
 | | Crystal structure of E. coli phosphoenolpyruvate carboxykinase mutant Lys254Ser | | Descriptor: | ACETATE ION, CALCIUM ION, Phosphoenolpyruvate carboxykinase (ATP) | | Authors: | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
1LAI
 
 | | Solution Structure of the B-DNA Duplex CGCGGTGTCCGCG. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*CP*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*GP*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-28 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
1L0W
 
 | | Aspartyl-tRNA synthetase-1 from space-grown crystals | | Descriptor: | Aspartyl-tRNA synthetase | | Authors: | Ng, J.D, Sauter, C, Lorber, B, Kirkland, N, Arnez, J, Giege, R. | | Deposit date: | 2002-02-14 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparative analysis of space-grown and earth-grown crystals of an aminoacyl-tRNA synthetase: space-grown crystals are more useful for structural determination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L1M
 
 | | SOLUTION STRUCTURE OF A DIMER OF LAC REPRESSOR DNA-BINDING DOMAIN COMPLEXED TO ITS NATURAL OPERATOR O1 | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3', Lactose operon repressor | | Authors: | Kalodimos, C.G, Bonvin, A.M.J.J, Salinas, R.K, Wechselberger, R, Boelens, R, Kaptein, R. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain.
EMBO J., 21, 2002
|
|
1L3Y
 
 | |
1LAR
 
 | | CRYSTAL STRUCTURE OF THE TANDEM PHOSPHATASE DOMAINS OF RPTP LAR | | Descriptor: | PROTEIN (LAR) | | Authors: | Nam, H.-J, Poy, F, Krueger, N, Saito, H, Frederick, C.A. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the tandem phosphatase domains of RPTP LAR.
Cell(Cambridge,Mass.), 97, 1999
|
|
1LBQ
 
 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
1KXZ
 
 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
8EIT
 
 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
