7AL8
 
 | |
3PE5
 
 | | Three-dimensional Structure of protein A7VV38_9CLOT from Clostridium leptum DSM 753, Northeast Structural Genomics Consortium Target QlR103 | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Northeast Structural Genomics Consortium Target QlR103
To be published
|
|
6ZO4
 
 | | The pointed end complex of dynactin bound to BICD2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
1IAZ
 
 | | EQUINATOXIN II | | Descriptor: | EQUINATOXIN II, SULFATE ION | | Authors: | Athanasiadis, A, Anderluh, G, Macek, P, Turk, D. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina.
Structure, 9, 2001
|
|
8K9L
 
 | | Full agonist- and positive allosteric modulator-bound mu-type opioid receptor-G protein complex | | Descriptor: | (2~{S})-2-(3-bromanyl-4-methoxy-phenyl)-3-(4-chlorophenyl)sulfonyl-1,3-thiazolidine, DAMGO, G protein subunit alpha i3, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
8K9K
 
 | | Full agonist-bound mu-type opioid receptor-G protein complex | | Descriptor: | DAMGO, G protein subunit alpha i3, Guanine nucleotide binding protein, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
1FNL
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF A HUMAN FCGRIII | | Descriptor: | LOW AFFINITY IMMUNOGLOBULIN GAMMA FC REGION RECEPTOR III-B, MERCURY (II) ION | | Authors: | Zhang, Y, Boesen, C.C, Radaev, S, Brooks, A.G, Fridman, W.H, Sautes-Fridman, C, Sun, P.D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the extracellular domain of a human Fc gamma RIII.
Immunity, 13, 2000
|
|
3QXF
 
 | |
3QRC
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
3QU3
 
 | | Crystal structure of IRF-7 DBD apo form | | Descriptor: | 1,2-ETHANEDIOL, Interferon regulatory factor 7, SODIUM ION | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
7A0S
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
3QXQ
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Zimmer, J. | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ.
J.Biol.Chem., 286, 2011
|
|
7A0R
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
8JEI
 
 | | Cryo-EM Structure of the compuond 5c-HCAR3-Gi complex | | Descriptor: | 3-nitro-4-(propylamino)benzoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
7A18
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin IV | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7ASN
 
 | |
1EJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
7ASM
 
 | |
8JEF
 
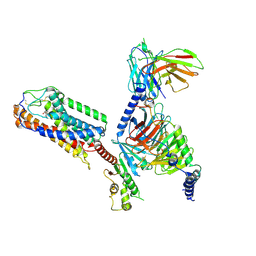 | | Cryo-EM Structure of the 3HO-HCAR3-Gi complex | | Descriptor: | (3S)-3-hydroxyoctanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
7AU6
 
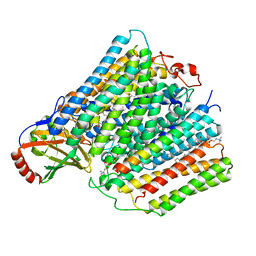 | |
7ATN
 
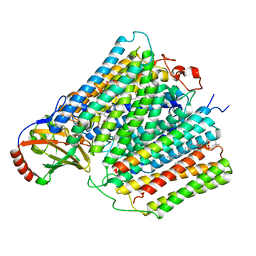 | |
7ATE
 
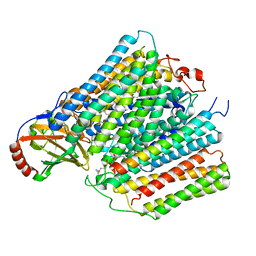 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
7AU3
 
 | | Cytochrome c oxidase structure in F-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cytochrome c oxidase structure in F-state
To Be Published
|
|
3NNY
 
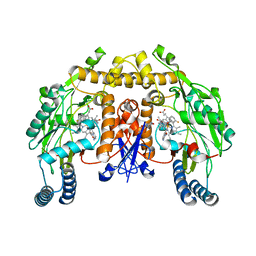 | | Structure of rat neuronal nitric oxide synthase heme domain complexed with 6-(((3R,4R)-4-(2-(3-Fluorophenethylamino)ethoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2010-06-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peripheral but crucial: a hydrophobic pocket (Tyr(706), Leu(337), and Met(336)) for potent and selective inhibition of neuronal nitric oxide synthase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
