2L7D
 
 | | Ribonucleotide Perturbation of DNA Structure: Solution Structure of [d(CGC)r(G)d(AATTCGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*G)-D(P*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | DeRose, E.F, Perera, L, Murray, M.S, Kunkel, T.A, London, R.E. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide.
Biochemistry, 51, 2012
|
|
426D
 
 | |
8VIU
 
 | |
1N0W
 
 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
5DAI
 
 | |
2NQH
 
 | |
2O8E
 
 | | human MutSalpha (MSH2/MSH6) bound to a G T mispair, with ADP bound to MSH2 only | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*CP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*GP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2O8B
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2LE2
 
 | | Novel dimeric structure of phage phi29-encoded protein p56: Insights into Uracil-DNA glycosylase inhibition | | Descriptor: | P56 | | Authors: | Asensio, J, Perez-Lago, L, Lazaro, J.M, Gonzalez, C, Serrano-Heras, G, Salas, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel dimeric structure of phage 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition.
Nucleic Acids Res., 39, 2011
|
|
2O8C
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and an O6-methyl-guanine T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*(6OG)P*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2O8D
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G dU mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*(DU)P*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
8G2N
 
 | |
7L6S
 
 | |
2IVK
 
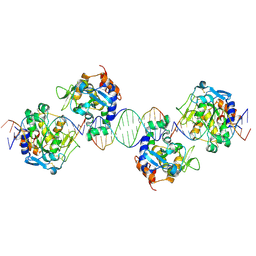 | | Crystal structure of the periplasmic endonuclease Vvn complexed with a 16-bp DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*CP*GP*AP*TP*CP*GP *AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*T)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*TP*C)-3', ... | | Authors: | Wang, Y.T, Yang, W.J, Li, C.L, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2006-06-14 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Sequence-Dependent DNA Cleavage by Nonspecific Endonucleases.
Nucleic Acids Res., 35, 2007
|
|
6RSO
 
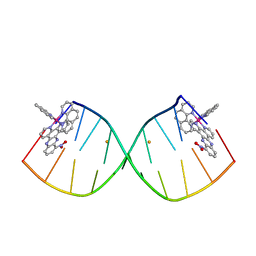 | |
5CTW
 
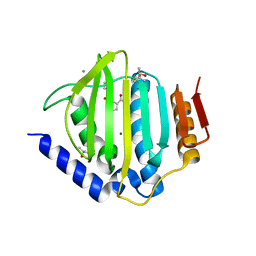 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
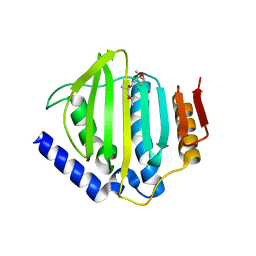 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
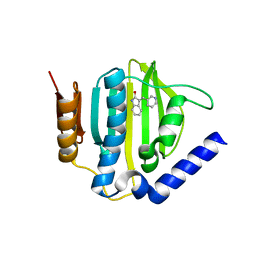 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1FN2
 
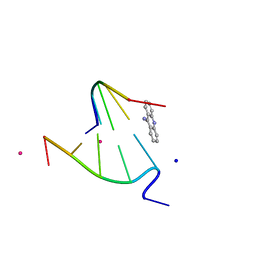 | | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1MUD
 
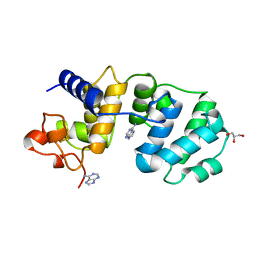 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | Descriptor: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
5NZW
 
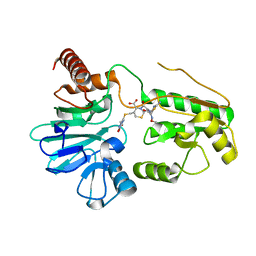 | | Crystal structure of DNA cross-link repair protein 1A in complex with ceftriaxone | | Descriptor: | Ceftriaxone, DNA cross-link repair 1A protein, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Kupinska, K, Burgess-Brown, N.A, Talon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DNA cross-link repair protein 1A in complex with ceftriaxone
To Be Published
|
|
5NZY
 
 | | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, DNA cross-link repair 1A protein, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Kupinska, K, Burgess-Brown, N.A, Talon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime
To Be Published
|
|
1SK5
 
 | |
3LZ8
 
 | | Structure of a putative chaperone dnaj from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 2.9 a resolution. | | Descriptor: | Putative chaperone DnaJ | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Bearden, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Putative Chaperone Dnaj from Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 at 2.9 A Resolution.
To be Published
|
|
