1EFS
 
 | | CONFORMATION OF A DNA-RNA HYBRID | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*UP*CP*UP*UP*CP*CP*UP*CP*UP*C)-3') | | Authors: | Larue, V, Hantz, E. | | Deposit date: | 2000-02-10 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of an RNA--DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand.
Int.J.Biol.Macromol., 28, 2001
|
|
4ED6
 
 | | Human DNA polymerase eta - DNA ternary complex: Reaction in the AT crystal at pH 6.7 for 15 hr, Sideway translocation | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*TP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Nakamura, T, Zhao, Y, Yang, W. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
3OQG
 
 | | Restriction endonuclease HPY188I in complex with substrate DNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(*GP*AP*TP*CP*TP*GP*AP*AP*C)-3', DNA 5'-D(*GP*TP*TP*CP*AP*GP*AP*TP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
1DRR
 
 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH DNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
6IUE
 
 | | DNA helical wire containing Hg(II) | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Ono, A, Kanazawa, H, Ito, H, Goto, M, Nakamura, K, Saneyoshi, H, Kondo, J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5YX2
 
 | |
7EAQ
 
 | |
3JXY
 
 | |
1RRD
 
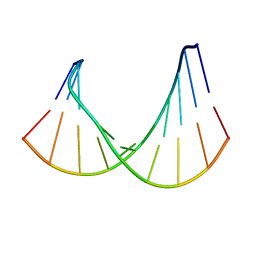 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH RNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
3JY1
 
 | |
3JXZ
 
 | |
2ZJT
 
 | |
1EI1
 
 | | DIMERIZATION OF E. COLI DNA GYRASE B PROVIDES A STRUCTURAL MECHANISM FOR ACTIVATING THE ATPASE CATALYTIC CENTER | | Descriptor: | DNA GYRASE B, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Brino, L, Urzhumtsev, A, Oudet, P, Moras, D. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center.
J.Biol.Chem., 275, 2000
|
|
5VZL
 
 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
1QZQ
 
 | | human Tyrosyl DNA phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase 1 | | Authors: | Raymond, A.C, Rideout, M.C, Staker, B, Hjerrild, K, Burgin Jr, A.B. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues.
J.Mol.Biol., 338, 2004
|
|
6AEQ
 
 | |
219D
 
 | |
1SUU
 
 | |
4H5A
 
 | |
1FJX
 
 | | STRUCTURE OF TERNARY COMPLEX OF HHAI METHYLTRANSFERASE MUTANT (T250G) IN COMPLEX WITH DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), HHAI DNA METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Vilkaitis, G, Dong, A, Weinhold, E, Cheng, X, Klimasauskas, S. | | Deposit date: | 2000-08-08 | | Release date: | 2000-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase.
J.Biol.Chem., 275, 2000
|
|
2QEG
 
 | | B-DNA with 7-deaza-dG modification | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DTP*DCP*(7GU)P*DCP*DG)-3') | | Authors: | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2007-06-25 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
2ADM
 
 | | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
1D0Q
 
 | |
1CY7
 
 | |
1CY4
 
 | |
