4GJY
 
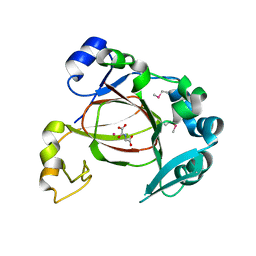 | | JMJD5 in complex with N-Oxalylglycine | | Descriptor: | COBALT (II) ION, JmjC domain-containing protein 5, N-OXALYLGLYCINE | | Authors: | Del Rizzo, P.A, Trievel, R.C. | | Deposit date: | 2012-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.2492 Å) | | Cite: | Crystal Structure and Functional Analysis of JMJD5 Indicate an Alternate Specificity and Function.
Mol.Cell.Biol., 32, 2012
|
|
4GK9
 
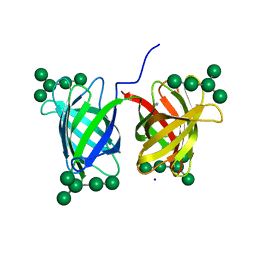 | | Crystal structure of Burkholderia oklahomensis agglutinin (BOA) bound to 3a,6a-mannopentaose | | Descriptor: | IMIDAZOLE, SODIUM ION, agglutinin (BOA), ... | | Authors: | Whitley, M.J, Furey, W, Gronenborn, A.M. | | Deposit date: | 2012-08-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity.
Febs J., 280, 2013
|
|
3E83
 
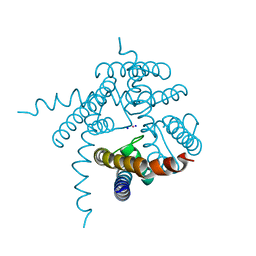 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
3S2G
 
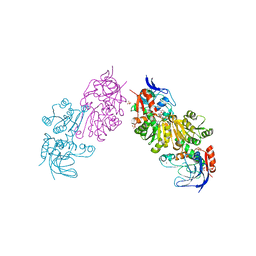 | | Crystal Structure of FurX NADH+:Furfuryl alcohol I | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
2OVG
 
 | | Lambda Cro Q27P/A29S/K32Q triple mutant at 1.35 A in space group P3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phage lambda Cro, SULFATE ION | | Authors: | Hall, B.M, Heroux, A, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
3E9C
 
 | |
4EG0
 
 | |
3EA3
 
 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
4GM2
 
 | | The crystal structure of a peptidase from plasmodium falciparum | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | El Bakkouri, M, Jung, P, Wernimont, A.K, Calmettes, C, Hui, R, Houry, W.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-15 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
3S4Y
 
 | | Crystal structure of human thiamin pyrophosphokinase 1 | | Descriptor: | CALCIUM ION, SULFATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Li, Y, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human thiamin pyrophosphokinase 1
to be published
|
|
4EHW
 
 | | Crystal structure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3S12
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, strain YU562 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2011-05-14 | | Release date: | 2011-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity.
Plos Pathog., 7, 2011
|
|
4EIN
 
 | | Crystal structure of mouse thymidylate synthase in binary complex with a substrate analogue and strong inhibitor, N(4)-hydroxy-2'-deoxycytidine-5'-monophosphate | | Descriptor: | 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), GLYCEROL, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Wilk, P, Kierdaszuk, B, Rode, W. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of complexes of mouse thymidylate synthase
crystallized with N4-OH-dCMP alone or in the presence of
N5,10-methylenetetrahydrofolate
PTERIDINES, 2013
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3S74
 
 | |
2OXM
 
 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
4GMY
 
 | |
3S9W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS M98G bound to Ca2+ and thymidine-5',3'-diphosphate at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2011-06-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS M98G bound to Ca2+ and thymidine-5',3'-diphosphate at cryogenic temperature
To be Published
|
|
4GMU
 
 | |
3S5P
 
 | |
2OZJ
 
 | |
4GNQ
 
 | |
3SC4
 
 | |
3FM9
 
 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
