1QNT
 
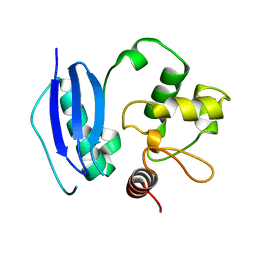 | |
1B4E
 
 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
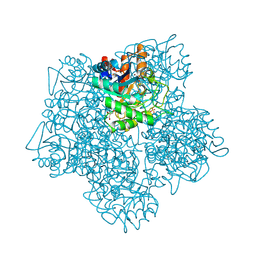
1B37
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (POLYAMINE OXIDASE), ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
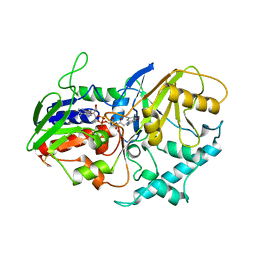
1C5G
 
 | | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Goldsmith, E.J. | | Deposit date: | 1999-12-07 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering of plasminogen activator inhibitor-1 to reduce the rate of latency transition.
Nat.Struct.Biol., 2, 1995
|
|
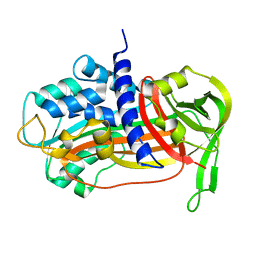
1DWM
 
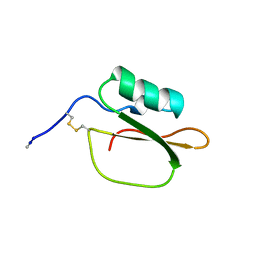 | |
2DLF
 
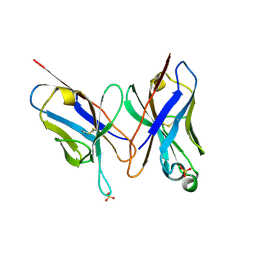 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
1QU5
 
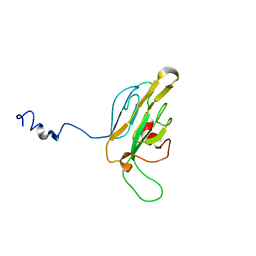 | |
3UBP
 
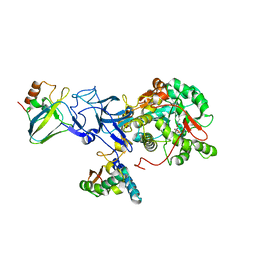 | | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | | Descriptor: | DIAMIDOPHOSPHATE, NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Miletti, S, Mangani, S, Ciurli, S. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
485D
 
 | |
432D
 
 | | D(GGCCAATTGG) COMPLEXED WITH DAPI | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3') | | Authors: | Vlieghe, D, Van Meervelt, L. | | Deposit date: | 1998-10-14 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of d(GGCCAATTGG) complexed with DAPI reveals novel binding mode.
Biochemistry, 38, 1999
|
|
1QRV
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HMG-D AND DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3'), HIGH MOBILITY GROUP PROTEIN D, SODIUM ION | | Authors: | Murphy IV, F.V, Sweet, R.M, Churchill, M.E.A. | | Deposit date: | 1999-06-15 | | Release date: | 1999-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition.
EMBO J., 18, 1999
|
|
1DGW
 
 | |
2CBR
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN I IN COMPLEX WITH A RETINOBENZOIC ACID (AM80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, PROTEIN (CRABP-I) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CW2
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-25 | | Release date: | 1999-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
2MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT F329I | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
1C3X
 
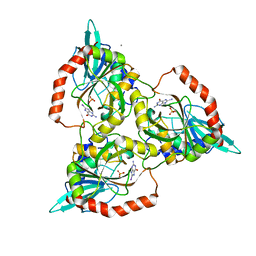 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH 8-IODO-GUANINE | | Descriptor: | 8-IODO-GUANINE, CALCIUM ION, PENTOSYLTRANSFERASE, ... | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-29 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
1WBF
 
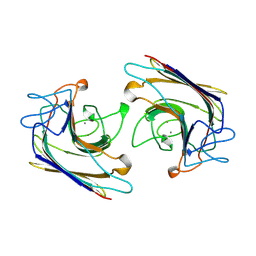 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1D3B
 
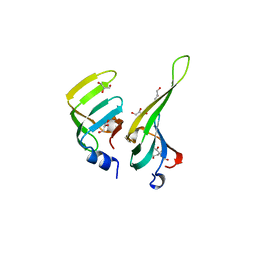 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | Authors: | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1GRN
 
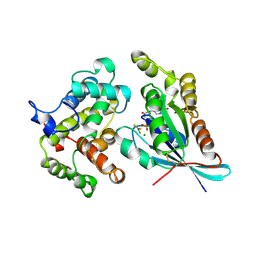 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1D2B
 
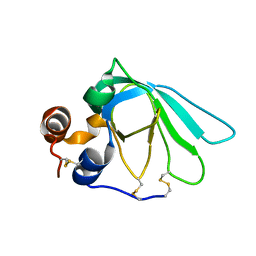 | | THE MMP-INHIBITORY, N-TERMINAL DOMAIN OF HUMAN TISSUE INHIBITOR OF METALLOPROTEINASES-1 (N-TIMP-1), SOLUTION NMR, 29 STRUCTURES | | Descriptor: | Metalloproteinase inhibitor 1 | | Authors: | Wu, B, Arumugam, S, Semenchenko, V, Brew, K, Van Doren, S.R. | | Deposit date: | 1999-09-22 | | Release date: | 1999-12-22 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of tissue inhibitor of metalloproteinases-1 implicates localized induced fit in recognition of matrix metalloproteinases.
J.Mol.Biol., 295, 2000
|
|
1CH0
 
 | | RNASE T1 VARIANT WITH ALTERED GUANINE BINDING SEGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, GUANOSINE-2'-MONOPHOSPHATE, ... | | Authors: | Hoeschler, K, Hoier, H, Orth, P, Hubner, B, Saenger, W, Hahn, U. | | Deposit date: | 1999-03-30 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of an RNase T1 variant with an altered guanine binding segment.
J.Mol.Biol., 294, 1999
|
|
1CR5
 
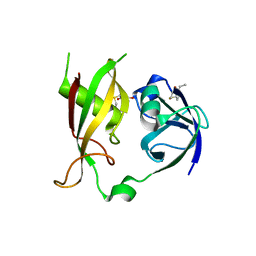 | | N-TERMINAL DOMAIN OF SEC18P | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, SEC18P (RESIDUES 22 - 210) | | Authors: | Babor, S.M, Fass, D. | | Deposit date: | 1999-08-13 | | Release date: | 1999-12-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Sec18p N-terminal domain.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D3C
 
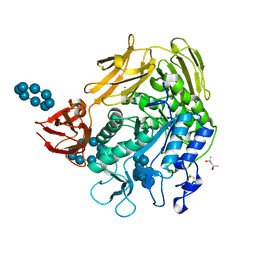 | | MICHAELIS COMPLEX OF BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE WITH GAMMA-CYCLODEXTRIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, van der Veen, B.A, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 1999-09-29 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a gamma-cyclodextrin-CGTase complex at 1.8-A resolution.
J.Biol.Chem., 274, 1999
|
|
3CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-12 7310) | | Descriptor: | (2E,4E,6E,8E)-9-(4-hydroxy-2,3,6-trimethylphenyl)-3,7-dimethylnona-2,4,6,8-tetraenoic acid, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
