5W4Q
 
 | |
5BSF
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5IWI
 
 | | 1.98A structure of GSK945237 with S.aureus DNA gyrase and singly nicked DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6N4M
 
 | | IDS-oxidized ADP-bound form of the nitrogenase Fe-protein from A. vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wenke, B.B, Spatzal, T, Rees, D.C. | | Deposit date: | 2018-11-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Site-Specific Oxidation State Assignments of the Iron Atoms in the [4Fe:4S]2+/1+/0States of the Nitrogenase Fe-Protein.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7ZKP
 
 | | Late assembly intermediate of the proximal proton pumping module of complex I with assembly factors NDUFAF1 and CIA84 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, ... | | Authors: | Schiller, J, Laube, E, Vonck, J, Zickermann, V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling.
Sci Adv, 8, 2022
|
|
5ZV3
 
 | |
5W4P
 
 | |
5VYL
 
 | |
5W4O
 
 | |
6PW1
 
 | | Cytochrome c Oxidase delta 16 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
1V4B
 
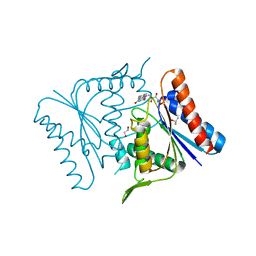 | |
5BSE
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5Z8Z
 
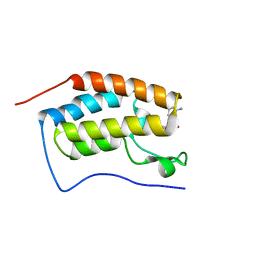 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 6-[(3S)-3-azanylpiperidin-1-yl]sulfonyl-1-ethyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
1AV6
 
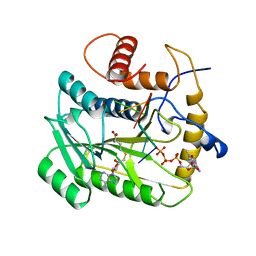 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M7G CAPPED RNA HEXAMER AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, RNA (5'-R(*GP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1997-09-26 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme.
Mol.Cell, 1, 1998
|
|
1PHB
 
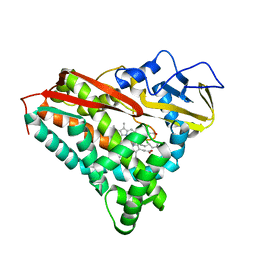 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
7L0H
 
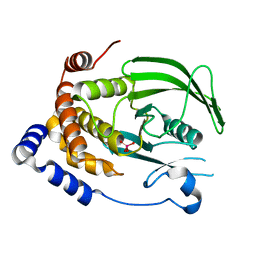 | | Vanadate-bound PTP1B T177G | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | Authors: | Shen, R.D, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
6T0B
 
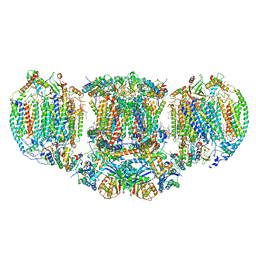 | | The III2-IV(5B)2 respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Marechal, A, Pinotsis, N, Hartley, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3KSZ
 
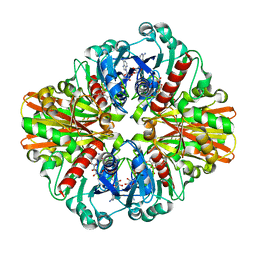 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
5J1E
 
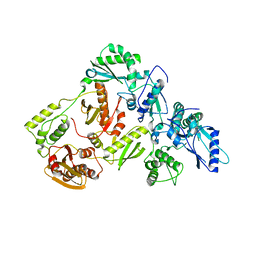 | | Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 5-hydroxy-4-oxo-1-[(4'-sulfamoyl[1,1'-biphenyl]-4-yl)methyl]-1,4-dihydropyridine-3-carboxylic acid, HIV-1 REVERSE TRANSCRIPTASE P51 DOMAIN, HIV-1 REVERSE TRANSCRIPTASE P66 DOMAIN, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H.
J.Med.Chem., 59, 2016
|
|
4JZ6
 
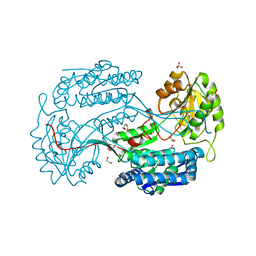 | | Crystal structure of a salicylaldehyde dehydrogenase from Pseudomonas putida G7 complexed with salicylaldehyde | | Descriptor: | 1,2-ETHANEDIOL, SALICYLALDEHYDE, SULFATE ION, ... | | Authors: | Coitinho, J.B, Nagem, R.A.P. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structural and Kinetic Properties of the Aldehyde Dehydrogenase NahF, a Broad Substrate Specificity Enzyme for Aldehyde Oxidation.
Biochemistry, 55, 2016
|
|
4Q3W
 
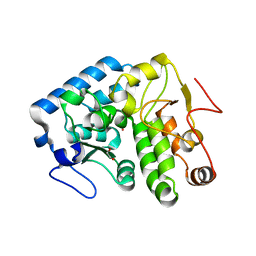 | | Crystal structure of C. violaceum phenylalanine hydroxylase D139E mutation | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2014-04-12 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved acidic residue in phenylalanine hydroxylase contributes to cofactor affinity and catalysis.
Biochemistry, 53, 2014
|
|
6A9W
 
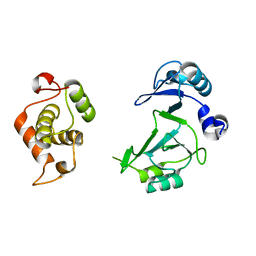 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
3KW4
 
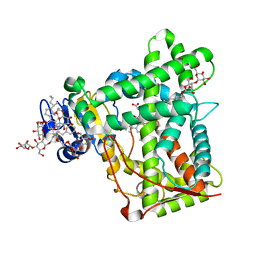 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
3KSD
 
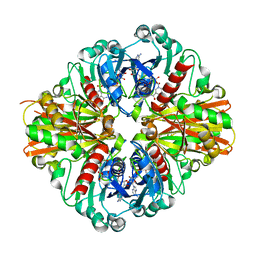 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.2 angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-22 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
6N4J
 
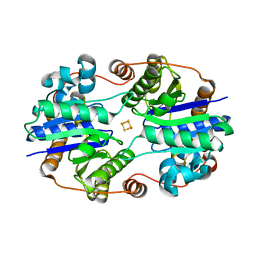 | |
