7UK1
 
 | |
7UD5
 
 | | Complex between MLL1-WRAD and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-03-18 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8QRN
 
 | | mt-SSU in GTPBP8 knock-out cells, state 4 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
4JLP
 
 | |
8QRM
 
 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QU1
 
 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QRL
 
 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QU5
 
 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
7LG8
 
 | | EGFR (T79M/V948R) in complex with naquotinib and an allosteric inhibitor | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{6-[4-(1-methylpiperidin-4-yl)phenyl]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2021-01-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors.
Nat Commun, 13, 2022
|
|
8QRK
 
 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 1 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.69 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
3MQI
 
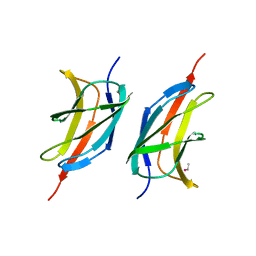 | | Human early B-cell factor 1 (EBF1) IPT/TIG domain | | Descriptor: | ETHYL MERCURY ION, Transcription factor COE1, trimethylamine oxide | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schueler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
7TZU
 
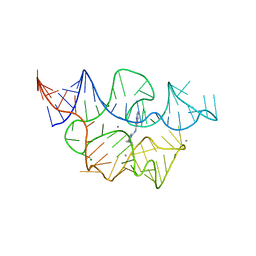 | |
4JL8
 
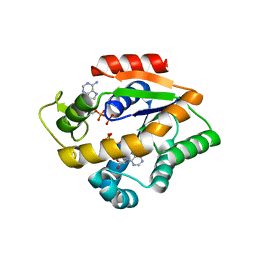 | |
7TZS
 
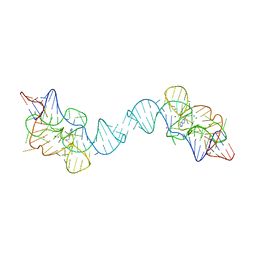 | |
4JLD
 
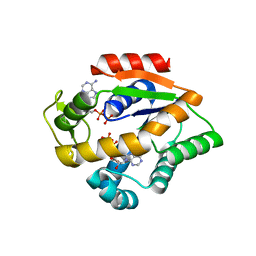 | |
7TZT
 
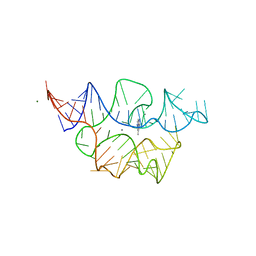 | | Crystal structure of the E. coli thiM riboswitch in complex with N1,N1-dimethyl-N2-(quinoxalin-6-ylmethyl)ethane-1,2-diamine (linked compound 37) | | Descriptor: | 6-methylquinoxaline, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | SHAPE-enabled fragment-based ligand discovery for RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TZR
 
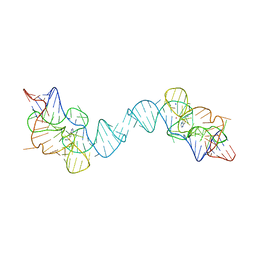 | |
4JLA
 
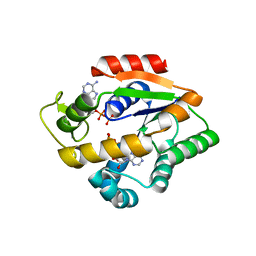 | |
6QKI
 
 | | Native structure of EgtB from Chloracidobacterium thermophilum, a type II sulfoxide synthase | | Descriptor: | FE (III) ION, Uncharacterized protein | | Authors: | Stampfli, A.R, Badri, B.N, Schirmer, T, Seebeck, F.P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An Alternative Active Site Architecture for O2Activation in the Ergothioneine Biosynthetic EgtB from Chloracidobacterium thermophilum.
J.Am.Chem.Soc., 141, 2019
|
|
3MUJ
 
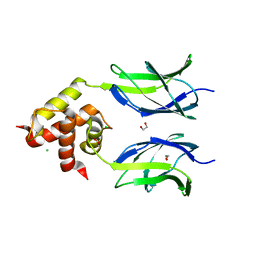 | | Early B-cell factor 3 (EBF3) IPT/TIG and dimerization helices | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Transcription factor COE3 | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schueler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
1UHL
 
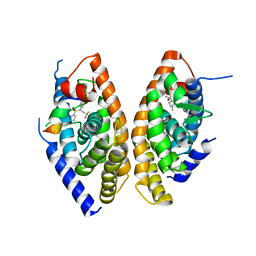 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
3N9V
 
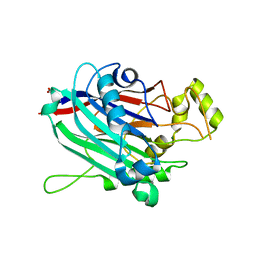 | | Crystal Structure of INPP5B | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
8E1G
 
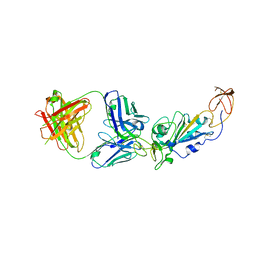 | | SARS-CoV-2 RBD in complex with Omicron-neutralizing antibody 2A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2A10 Fab, heavy chain, ... | | Authors: | Wasserman, H, Hastie, K.M, Buck, T.K, Saphire, E.O. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
4ACX
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 23 | | Descriptor: | (8R)-8-[4-(DIFLUOROMETHOXY)PHENYL]-3,3-DIFLUORO-8-[3-(3-METHOXYPROP-1-YN-1-YL)PHENYL]-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
