8XXB
 
 | |
8PT0
 
 | | ERK2 covelently bound to RU75 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
8XPE
 
 | | Crystal structure of Tris-bound TsaBgl (DATA III) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8PV0
 
 | | Crystal structure of tropomyosin (Cdc8) cables, Conformers 2 and 3 | | Descriptor: | Tropomyosin | | Authors: | Reinke, P.Y.A, Zahn, M, Fedorov, R, Manstein, D.J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-24 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structures of cables formed by the acetylated and unacetylated forms of the Schizosaccharomyces pombe tropomyosin ortholog Tpm Cdc8.
J.Biol.Chem., 300, 2024
|
|
8XX9
 
 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus.
Proteins, 92, 2024
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
7SLR
 
 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
8XU5
 
 | |
8PKR
 
 | |
8XPC
 
 | | Crystal structure of Tris-bound TsaBgl (DATA I) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
7SFN
 
 | |
8XPD
 
 | | Crystal structure of Tris-bound TsaBgl (DATA II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
7SEI
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403Q) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
8YA5
 
 | |
6CKR
 
 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | Bromodomain-containing protein 4, N-{3-[2-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1-oxo-1,2-dihydroisoquinolin-4-yl]phenyl}methanesulfonamide | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8YA1
 
 | | HEN EGG WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8YBG
 
 | |
6CL2
 
 | |
8Y4U
 
 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
8YA4
 
 | |
8PKB
 
 | | Staphylococcus aureus endonuclease IV with bound phosphate | | Descriptor: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Saper, M, Kirillov, S, Rouvinski, A. | | Deposit date: | 2023-06-26 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli .
Biochemistry, 64, 2025
|
|
6D12
 
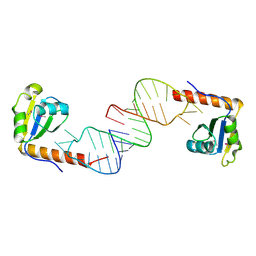 | | Crystal structure of C-terminal xRRM domain of human Larp7 bound to 7SK stem-loop 4 RNA | | Descriptor: | La-related protein 7, human 7SK RNA stem-loop 4 | | Authors: | Eichhorn, C.D, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8YBH
 
 | |
