5C1F
 
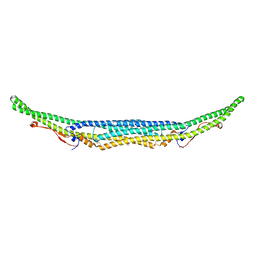 | | Structure of the Imp2 F-BAR domain | | Descriptor: | FORMIC ACID, Septation protein imp2 | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3551 Å) | | Cite: | The Tubulation Activity of a Fission Yeast F-BAR Protein Is Dispensable for Its Function in Cytokinesis.
Cell Rep, 14, 2016
|
|
1CSK
 
 | |
1IOZ
 
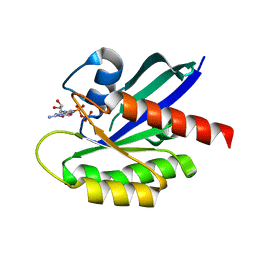 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1IOU
 
 | |
5CJ2
 
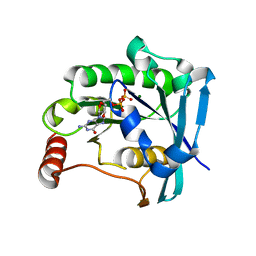 | | Ran GDP Y39A mutant triclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CHX
 
 | | Crystal Structure of amino acids 1590-1657 of MYH7 | | Descriptor: | Xrcc4-MYH7-1590-1657 | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinz, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-10 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
5CLQ
 
 | | Ran Y39A in complex with GPPNHP and RanBD1 | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
1J4E
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE COVALENTLY BOUND TO THE SUBSTRATE DIHYDROXYACETONE PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE A | | Authors: | Choi, K.H, Shi, J, Hopkins, C.E, Tolan, D.R, Allen, K.N. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry, 40, 2001
|
|
5JLE
 
 | | Crystal structure of SETD2 bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, H, Yang, S, Zheng, X. | | Deposit date: | 2016-04-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
1IWQ
 
 | | Crystal Structure of MARCKS calmodulin binding domain peptide complexed with Ca2+/Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, MARCKS | | Authors: | Yamauchi, E, Nakatsu, T, Matsubara, M, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-31 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca(2+)-calmodulin
NAT.STRUCT.BIOL., 10, 2003
|
|
1DFJ
 
 | |
1DFL
 
 | | SCALLOP MYOSIN S1 COMPLEXED WITH MGADP:VANADATE-TRANSITION STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Houdusse, A, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 1999-11-19 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Three conformational states of scallop myosin S1.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ISM
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with nicotinamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISG
 
 | | Crystal Structure Analysis of BST-1/CD157 with ATPgammaS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISN
 
 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
1DFK
 
 | |
1ISF
 
 | | Crystal Structure Analysis of BST-1/CD157 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
5CJ0
 
 | | Crystal Structure of Amino Acids 1631-1692 of MYH7 | | Descriptor: | Xrcc4-MYH7-(1631-1692) chimera protein | | Authors: | Korkmaz, N.E, Taylor, K.C, Andreas, M.P, Ajay, G, Heinze, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
1IFQ
 
 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
7KYZ
 
 | |
5CJ1
 
 | | Crystal structure of the coiled coil of MYH7 residues 1526 to 1571 fused to Gp7 | | Descriptor: | Gp7-MYH7-(1526-1571) chimera protein | | Authors: | Taylor, K.C, Korkmaz, E.N, Andreas, M.P, Ajay, G, Heinz, N.T, Cui, Q, Rayment, I. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A composite approach towards a complete model of the myosin rod.
Proteins, 84, 2016
|
|
1J19
 
 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | Descriptor: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | Authors: | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
5CIW
 
 | | Ran GDP Y39A mutant monoclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CIQ
 
 | | Ran GDP wild type tetragonal crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CIT
 
 | | Ran GDP wild type monoclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
