2HRK
 
 | |
6W45
 
 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
2HSN
 
 | | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes | | Descriptor: | GU4 nucleic-binding protein 1, Methionyl-tRNA synthetase, cytoplasmic | | Authors: | Simader, H, Koehler, C, Basquin, J, Suck, D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.
Nucleic Acids Res., 34, 2006
|
|
2HSM
 
 | |
3CCT
 
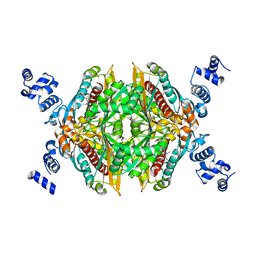 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-[(2-hydroxyphenyl)carbamoyl]-5-(1-methylethyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CCZ
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[2-(4-fluorophenyl)-4-{[(1S)-2-hydroxy-1-phenylethyl]carbamoyl}-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CD5
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(biphenyl-4-ylcarbamoyl)-2-ethyl-5,6,7,8-tetrahydrocyclohepta[b]pyrrol-1(4H)-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
8BN3
 
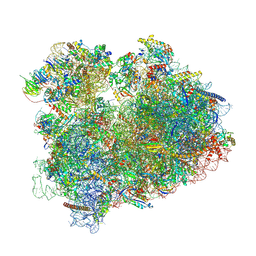 | | Yeast 80S, ES7s delta, eIF5A, Stm1 containing | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Dimitrova-Paternoga, L, Paternoga, H, Wilson, D.N. | | Deposit date: | 2022-11-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Evolving precision: rRNA expansion segment 7S modulates translation velocity and accuracy in eukaryal ribosomes.
Nucleic Acids Res., 52, 2024
|
|
1SMR
 
 | |
4KXT
 
 | |
4CXH
 
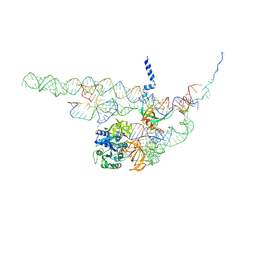 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S RRNA - H44, 18S RRNA - H5-H14, 18S RRNA - H8, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-07 | | Release date: | 2014-07-16 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
4CXG
 
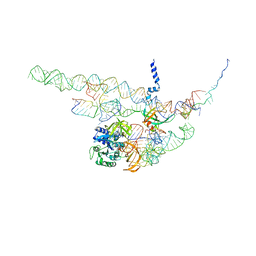 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S RRNA - H44, 18S RRNA - H5-H14, 18S RRNA - H8, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-07 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
3KTQ
 
 | |
4CSC
 
 | |
4V7R
 
 | | Yeast 80S ribosome. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Ben-Shem, A, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2010-07-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the eukaryotic ribosome.
Science, 330, 2010
|
|
2FTP
 
 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
5E2S
 
 | |
5E28
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(4-aminophenyl)benzenesulfonamide inhibitor | | Descriptor: | 4'-aminobiphenyl-4-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
J.Med.Chem., 59, 2016
|
|
2J3J
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3K
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
3FX4
 
 | | Porcine aldehyde reductase in ternary complex with inhibitor | | Descriptor: | Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with a 5-arylidene-2,4-thiazolidinedione aldose reductase inhibitor.
Eur.J.Med.Chem., 45, 2010
|
|
4L4S
 
 | | Structural characterisation of the NADH binary complex of human lactate dehydrogenase M isozyme | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6CTS
 
 | |
3JYC
 
 | |
