2LKK
 
 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
1SOL
 
 | | A PIP2 AND F-ACTIN-BINDING SITE OF GELSOLIN, RESIDUE 150-169 (NMR, AVERAGED STRUCTURE) | | Descriptor: | GELSOLIN (150-169) | | Authors: | Xian, W, Vegners, R, Janmey, P.A, Braunlin, W.H. | | Deposit date: | 1995-09-29 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles.
Biophys.J., 69, 1995
|
|
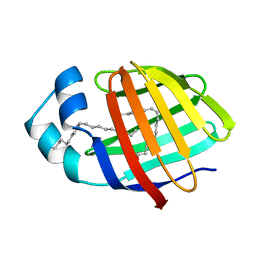
1HJI
 
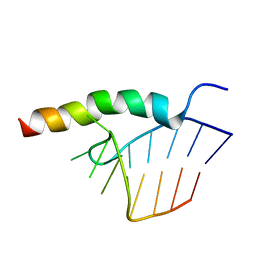 | | BACTERIOPHAGE HK022 NUN-PROTEIN-NUTBOXB-RNA COMPLEX | | Descriptor: | NUN-PROTEIN, RNA (5-R(P*GP*CP*CP*CP*UP*GP*AP*AP*AP*AP*AP*GP*GP*GP*C)-3) | | Authors: | Faber, C, Schaerpf, M, Becker, T, Sticht, H, Roesch, P. | | Deposit date: | 2001-01-15 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Coliphage Hk022 Nun Protein-Lambda-Phage Boxb RNA Complex. Implications for the Mechanism of Transcription Termination
J.Biol.Chem., 276, 2001
|
|
1I3Y
 
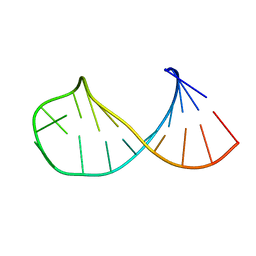 | |
1QZE
 
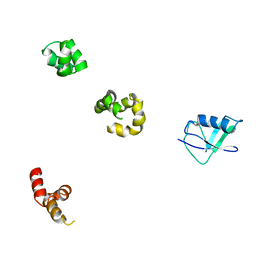 | | HHR23a protein structure based on residual dipolar coupling data | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Walters, K.J, Lech, P.J, Goh, A.M, Wang, Q, Howley, P.M. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R4D
 
 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
1J7Q
 
 | | Solution structure and backbone dynamics of the defunct EF-hand domain of Calcium Vector Protein | | Descriptor: | Calcium Vector Protein | | Authors: | Theret, I, Baladi, S, Cox, J.A, Gallay, J, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2001-05-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the defunct domain of calcium vector protein.
Biochemistry, 40, 2001
|
|
1IO6
 
 | |
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1IMW
 
 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1IT5
 
 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
1RJI
 
 | | Solution Structure of BmKX, a novel potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | potassium channel toxin KX | | Authors: | Cai, Z, Wu, J, Xu, Y, Wang, C.-G, Chi, C.-W, Shi, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A novel short-chain peptide BmKX from the Chinese scorpion Buthus martensi karsch, sequencing, gene cloning and structure determination
Toxicon, 45, 2005
|
|
1K4U
 
 | | Solution structure of the C-terminal SH3 domain of p67phox complexed with the C-terminal tail region of p47phox | | Descriptor: | PHAGOCYTE NADPH OXIDASE SUBUNIT P47PHOX, PHAGOCYTE NADPH OXIDASE SUBUNIT P67PHOX | | Authors: | Kami, K, Takeya, R, Sumimoto, H, Kohda, D. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
EMBO J., 21, 2002
|
|
1K42
 
 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1HUE
 
 | | HISTONE-LIKE PROTEIN | | Descriptor: | HU PROTEIN | | Authors: | Vis, H, Mariani, M, Vorgias, C.E, Wilson, K.S, Kaptein, R, Boelens, R. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HU protein from Bacillus stearothermophilus.
J.Mol.Biol., 254, 1995
|
|
1T6W
 
 | | RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, hypothetical protein XP_346638 | | Authors: | Yang, W, Wilkins, A.L, Ye, Y, Liu, Z.-R, Urbauer, J.L, Kearney, A, van der Merwe, P.A, Yang, J.J. | | Deposit date: | 2004-05-07 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design of a calcium-binding protein with desired structure in a cell adhesion molecule.
J.Am.Chem.Soc., 127, 2005
|
|
1TTF
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
1TTG
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
1HS2
 
 | |
1S37
 
 | | Accomodation of Mispair-Aligned N3T-Ethyl-N3T DNA Interstrand Crosslink | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*(TTM)P*TP*TP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*AP*AP*TP*TP*TP*TP*CP*G)-3') | | Authors: | da Silva, M.W, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Accommodation of mispair aligned N3T-ethyl-N3T DNA interstrand cross link.
Biochemistry, 43, 2004
|
|
1KX2
 
 | | Minimized average structure of a mono-heme ferrocytochrome c from Shewanella putrefaciens | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-30 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1RGW
 
 | | Solution Structure of ZASP's PDZ domain | | Descriptor: | ZASP protein | | Authors: | Au, Y, Atkinson, R.A, Pallavicini, A, Joseph, C, Martin, S.R, Muskett, F.W, Guerrini, R, Faulkner, G, Pastore, A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Structure, 12, 2004
|
|
1KVG
 
 | | EPO-3 beta Hairpin Peptide | | Descriptor: | Protein: EPO-3 Receptor Agonist | | Authors: | Skelton, N.J, Russell, S, de Sauvage, F, Cochran, A.G. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Determinants of beta-Hairpin Conformation in Erythropoeitin
Receptor Agonist Peptides Derived from a Phage Display Library
J.Mol.Biol., 316, 2002
|
|
1K8S
 
 | | BULGED ADENOSINE IN AN RNA DUPLEX | | Descriptor: | 5'-R(*GP*CP*GP*GP*CP*AP*CP*CP*UP*GP*CP*C)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*UP*GP*CP*CP*GP*C)-3' | | Authors: | Thiviyanathan, V, Guliaev, A.B, Leontis, N.B, Gorenstein, D.G. | | Deposit date: | 2001-10-25 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a bulged adenosine base in an RNA duplex by relaxation matrix refinement.
J.Mol.Biol., 300, 2000
|
|
1KBE
 
 | | Solution structure of the cysteine-rich C1 domain of Kinase Suppressor of Ras | | Descriptor: | Kinase Suppressor of Ras, ZINC ION | | Authors: | Zhou, M, Horita, D.A, Waugh, D.S, Byrd, R.A, Morrison, D.K. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR).
J.Mol.Biol., 315, 2002
|
|
