4R66
 
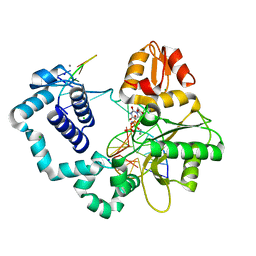 | | Ternary complex crystal structure of E295K mutant of DNA polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
3FB3
 
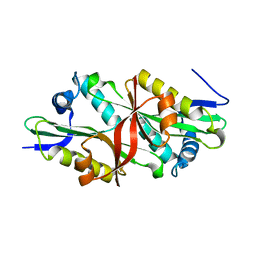 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3FOO
 
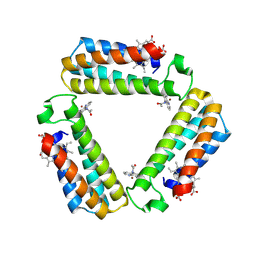 | |
4QT6
 
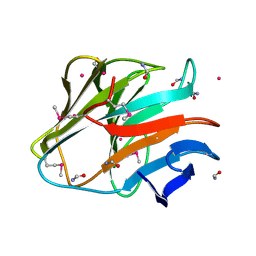 | | Crystal structure of the SPRY domain of human HERC1 | | Descriptor: | FORMAMIDE, Probable E3 ubiquitin-protein ligase HERC1, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Guan, X, Wernimont, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2015-01-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the SPRY domain of human HERC1
To be Published
|
|
3W62
 
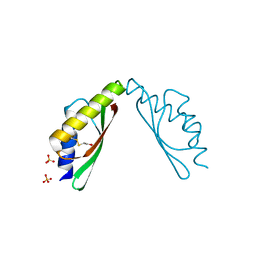 | | MamM-CTD E289A | | Descriptor: | Magnetosome protein MamM, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain
Plos One, 9, 2014
|
|
3W6B
 
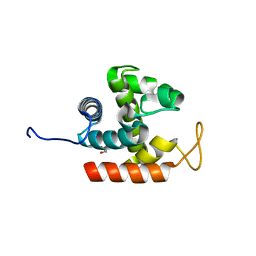 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 | | Descriptor: | GLYCEROL, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3FD4
 
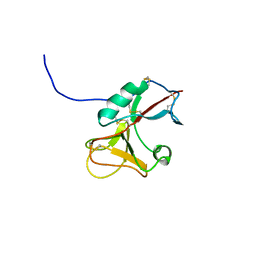 | |
3WEM
 
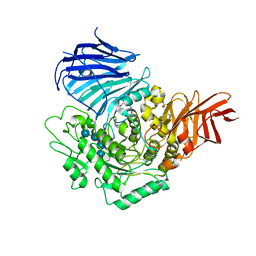 | | Sugar beet alpha-glucosidase with acarviosyl-maltotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3FD7
 
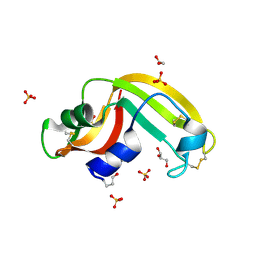 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
3EXB
 
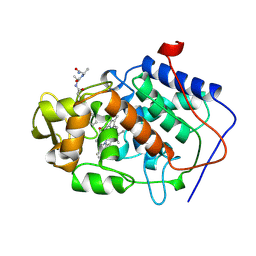 | |
3W7V
 
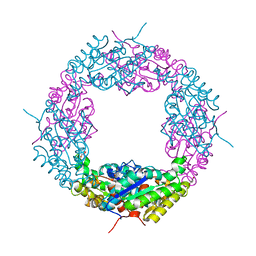 | | Crystal Structure of Axe2, an Acetylxylan Esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, CHLORIDE ION, GLYCEROL | | Authors: | Lansky, S, Alalouf, O, Solomon, H.V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WGJ
 
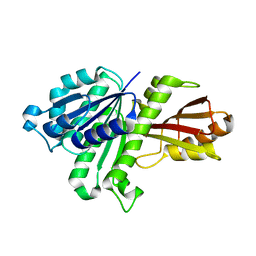 | |
3W8G
 
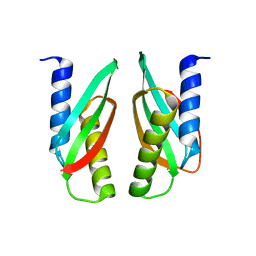 | | MamM V260R | | Descriptor: | Magnetosome protein MamM | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-03-12 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bacterial Magnetosome Biomineralization - A Novel Platform to Study Molecular Mechanisms of Human CDF-Related Type-II Diabetes
Plos One, 9, 2014
|
|
3WHB
 
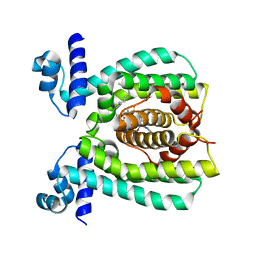 | |
3FG6
 
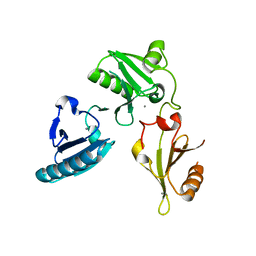 | | Structure of the C-terminus of Adseverin | | Descriptor: | Adseverin, CALCIUM ION | | Authors: | Robinson, R.C. | | Deposit date: | 2008-12-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the C-terminus of adseverin reveals the actin-binding interface.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3WIH
 
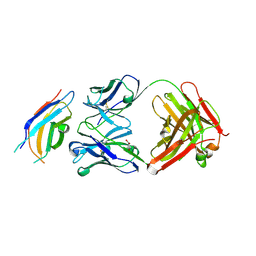 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3FM8
 
 | | Crystal structure of full length centaurin alpha-1 bound with the FHA domain of KIF13B (CAPRI target) | | Descriptor: | Centaurin-alpha-1, Kinesin-like protein KIF13B, SULFATE ION, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3FCF
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-[(1E,7E)-8-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,6-dioxa-2,7-diazaocta-1,7-dien-1-yl]benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3W5E
 
 | | Crystal structure of phosphodiesterase 4B in complex with compound 31e | | Descriptor: | CALCIUM ION, N-tert-butyl-2-{4-[(5,5-dioxido-2-phenyl-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)amino]phenyl}acetamide, ZINC ION, ... | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the fused bicyclic 4-amino-2-phenylpyrimidine derivatives as novel and potent PDE4 inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3WKS
 
 | | Crystal structure of the SepCysS-SepCysE N-terminal domain complex from | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.029 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3W6M
 
 | |
3CVR
 
 | | Crystal structure of the full length IpaH3 | | Descriptor: | Invasion plasmid antigen | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2008-04-19 | | Release date: | 2008-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Shigella effector reveals a new class of ubiquitin ligases
Nat.Struct.Mol.Biol., 15, 2008
|
|
3WSF
 
 | |
3CGN
 
 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
