7OKA
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 14 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-(1~{H}-imidazol-4-ylmethyl)ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
2G3O
 
 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T4I
 
 | | Crystal Structure of wild type EGFR in complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Skene, R.J, Lane, W. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
5AFS
 
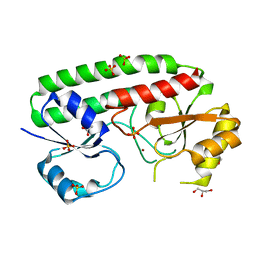 | | structure of Zn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis in Zn(2+)-bound state and biophysical characterization of CLas-ZnuA2.
Biochim. Biophys. Acta, 1864, 2016
|
|
4Y8F
 
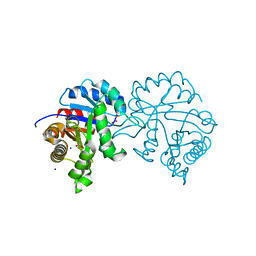 | |
4Y96
 
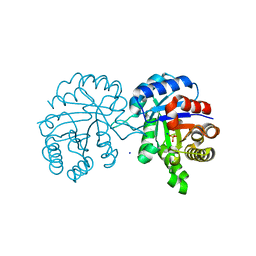 | | Crystal structure of Triosephosphate Isomerase from Gemmata obscuriglobus | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
4Y90
 
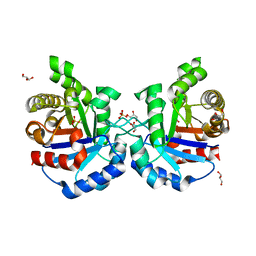 | | Crystal structure of Triosephosphate Isomerase from Deinococcus radiodurans | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernadez-Velasco, D.A. | | Deposit date: | 2015-02-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
4Y9A
 
 | |
5GVY
 
 | | Crystal structure of SALT protein from Oryza sativa | | Descriptor: | Salt stress-induced protein, alpha-D-mannopyranose | | Authors: | Sharma, P, Sagar, A, Kaur, N, Sharma, I, Kirat, K, Ashish, F.N.U, Pati, P.K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Structural insights into rice SalTol QTL located SALT protein.
Sci Rep, 10, 2020
|
|
2IRX
 
 | |
1LVY
 
 | | PORCINE ELASTASE | | Descriptor: | CALCIUM ION, ELASTASE, SULFATE ION | | Authors: | Schiltz, M, Prange, T. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-pressure krypton gas and statistical heavy-atom refinement: a successful combination of tools for macromolecular structure determination.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
5J5D
 
 | |
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
3CY6
 
 | | Crystal Structure of E18Q DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
2CMY
 
 | | Crystal complex between bovine trypsin and Veronica hederifolia trypsin inhibitor | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, GLYCEROL, ... | | Authors: | Conners, R, Yardley, J.L, Konarev, A, Shewry, P, Brady, R.L. | | Deposit date: | 2006-05-16 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An Unusual Helix-Turn-Helix Protease Inhibitory Motif in a Novel Trypsin Inhibitor from Seeds of Veronica (Veronica Hederifolia L.).
J.Biol.Chem., 282, 2007
|
|
4EZV
 
 | |
