2ADG
 
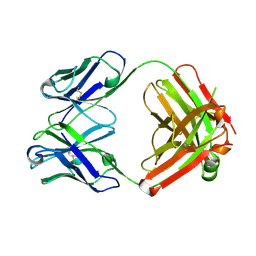 | | Crystal structure of monoclonal anti-CD4 antibody Q425 | | Descriptor: | Q425 Fab Light chain, Q425 Fab heavy chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2KRS
 
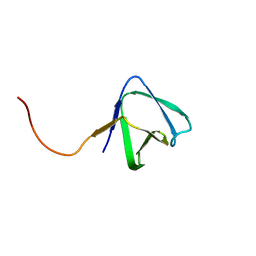 | | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. | | Descriptor: | Probable enterotoxin | | Authors: | Ramelot, T.A, Cort, J.R, Maglaqui, M, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
To be Published
|
|
3CVD
 
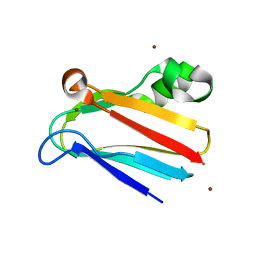 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (I) ION, Plastocyanin, ZINC ION | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
2ADJ
 
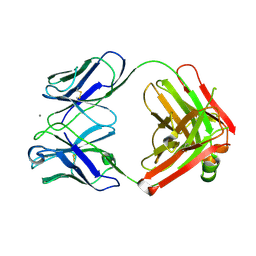 | | Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Calcium | | Descriptor: | CALCIUM ION, Q425 Fab Heavy chain, Q425 Fab Light chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3TTJ
 
 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3PTW
 
 | | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens Atcc 13124 | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Seidel, R, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens
Atcc 13124
To be Published
|
|
1N41
 
 | | Crystal Structure of Annexin V K27E Mutant | | Descriptor: | CALCIUM ION, SULFATE ION, annexin V | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial basic cluster in annexin V couples phospholipid
binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
4Q32
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(naphthalen-2-yl)-2-[2-(pyridin-2-yl)-1H-benzimidazol-1-yl]acetamide | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91
To be Published
|
|
4Q33
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110 | | Descriptor: | 4-[(1R)-1-[1-(4-chlorophenyl)-1,2,3-triazol-4-yl]ethoxy]-1-oxidanyl-quinoline, ACETIC ACID, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110
TO BE PUBLISHED
|
|
2JNK
 
 | |
1N42
 
 | | Crystal Structure of Annexin V R149E Mutant | | Descriptor: | Annexin V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Campos, B, Mealy, T.R, Commodore, L, Head, J.F, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial basic cluster in annexin V couples phospholipid binding and trimer formation on membrane surfaces
J.Biol.Chem., 278, 2003
|
|
3GFO
 
 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | Descriptor: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
4EDP
 
 | | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124 | | Descriptor: | ABC transporter, substrate-binding protein, ACETATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom resolution crystal structure of an ABC transporter from Clostridium perfringens ATCC 13124
To be Published
|
|
3UB1
 
 | | Ntf2 like protein involved in plasmid conjugation | | Descriptor: | DI(HYDROXYETHYL)ETHER, ORF13-like protein | | Authors: | Porter, C.J, Rosado, C.J, Bantwal, R, Bannam, T.L, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria.
Mol.Microbiol., 83, 2012
|
|
1EIN
 
 | |
3FCM
 
 | |
2JOV
 
 | | NMR Structure of Clostridium Perfringens Protein CPE0013. Northeast Structural Genomics Target CpR31. | | Descriptor: | Hypothetical protein CPE0013 | | Authors: | Ding, K, Ramelot, T.A, Anklin, C.G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-04 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Clostridium perfringens protein CPE0013.
To be Published
|
|
3F6A
 
 | |
2AN9
 
 | | Crystal Structure Of Oligomeric E.coli Guanylate Kinase In Complex With GDP | | Descriptor: | GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, Guanylate kinase, ... | | Authors: | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
2D1K
 
 | | Ternary complex of the WH2 domain of mim with actin-dnase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
2ANC
 
 | | Crystal Structure Of Unliganded Form Of Oligomeric E.coli Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
2EWH
 
 | | Carboxysome protein CsoS1A from Halothiobacillus neapolitanus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Major carboxysome shell protein 1A | | Authors: | Tsai, Y, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome.
Plos Biol., 5, 2007
|
|
2D1L
 
 | | Structure of F-actin binding domain IMD of MIM (Missing In Metastasis) | | Descriptor: | Metastasis suppressor protein 1 | | Authors: | Lee, S.H, Kerff, F, Chereau, D, Ferron, F, Dominguez, R. | | Deposit date: | 2005-08-27 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
1R8S
 
 | | ARF1[DELTA1-17]-GDP IN COMPLEX WITH A SEC7 DOMAIN CARRYING THE MUTATION OF THE CATALYTIC GLUTAMATE TO LYSINE | | Descriptor: | ADP-ribosylation factor 1, Arno, BETA-MERCAPTOETHANOL, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor
Nature, 426, 2003
|
|
1S9D
 
 | | ARF1[DELTA 1-17]-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-Ribosylation Factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2004-02-04 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor
Nature, 426, 2003
|
|
