1KF4
 
 | | Atomic Resolution Structure of RNase A at pH 6.3 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3NKK
 
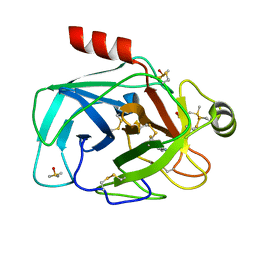 | | Trypsin in complex with fluorine containing fragment | | Descriptor: | 3-fluoro-4-methylbenzenecarboximidamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, Vulpetti, A, Dalvit, C. | | Deposit date: | 2010-06-20 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Combined use of computational chemistry, NMR screening, and X-ray crystallography for identification and characterization of fluorophilic protein environments.
Proteins, 78, 2010
|
|
3MTH
 
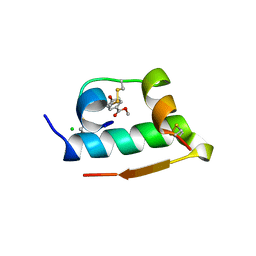 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
3NNN
 
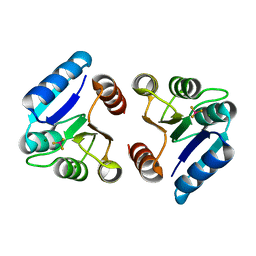 | | BeF3 Activated DrrD Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR D, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
1KT4
 
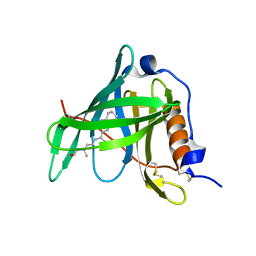 | | Crystal structure of bovine holo-RBP at pH 3.0 | | Descriptor: | RETINOL, plasma retinol-binding protein | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
3NSH
 
 | | BACE-1 in complex with ELN475957 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-3-({1-[4-(2,2-dimethylpropyl)thiophen-2-yl]cyclopropyl}amino)-2-hydroxypropyl]acetamide | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Brecht, E, Yao, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of hydroxyethylamine (HEA) BACE-1 inhibitors: structure-activity relationship of the aryl region.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1KE9
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
3NUP
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(1-methylpiperidin-4-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
3NUX
 
 | | CDK6 (monomeric) in complex with inhibitor | | Descriptor: | 4-[5-chloro-3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(5-piperazin-1-ylpyridin-2-yl)pyrimidin-2-amine, Cell division protein kinase 6 | | Authors: | Chopra, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6.
J.Med.Chem., 53, 2010
|
|
3NU3
 
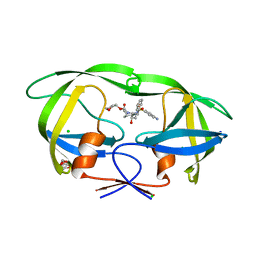 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
1KWY
 
 | | Rat mannose protein A complexed with man-a13-man. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
3NV6
 
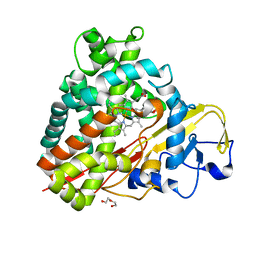 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
1KF7
 
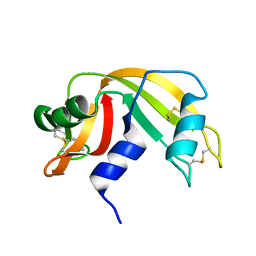 | | Atomic Resolution Structure of RNase A at pH 8.0 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3NWQ
 
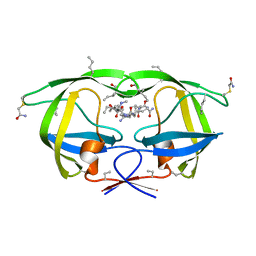 | |
1KE7
 
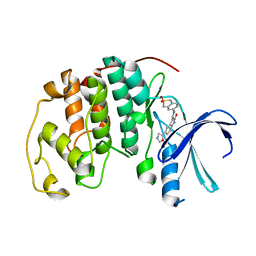 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
3O17
 
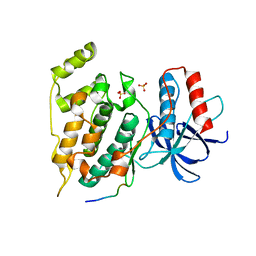 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
3O4M
 
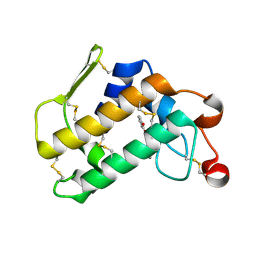 | | Crystal structure of porcine pancreatic phospholipase A2 in complex with 1,2-dihydroxybenzene | | Descriptor: | CALCIUM ION, CATECHOL, Phospholipase A2, ... | | Authors: | Dileep, K.V, Tintu, I, Karthe, P, Mandal, P.K, Haridas, M, Sadasivan, C. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to PLA(2) may contribute to the anti-inflammatory activity of catechol
Chem.Biol.Drug Des., 2011
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NPL
 
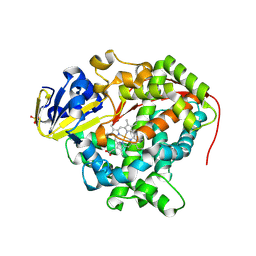 | | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Ener, M, Lee, Y.-T, Goodin, D.B, Winkler, J.R, Gray, H.B, Cheruzel, L. | | Deposit date: | 2010-06-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Ru(bpy)2(A-Phen)(K97C) P450 BM3 heme domain, a ruthenium modified P450 BM3 mutant
To be Published
|
|
3NWW
 
 | | P38 Alpha kinase complexed with a 2-aminothiazol-5-yl-pyrimidine based inhibitor | | Descriptor: | 1-[2-(2-{[2-(dimethylamino)ethyl]amino}-6-{2-[(1-methylethyl)amino]-1,3-thiazol-5-yl}pyrimidin-4-yl)benzyl]-3-ethylurea, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Utilization of a nitrogen-sulfur nonbonding interaction in the design of new 2-aminothiazol-5-yl-pyrimidines as p38alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O20
 
 | | Electron transfer complexes:experimental mapping of the Redox-dependent Cytochrome C electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Casini, A, Messori, L, Geremia, S, Demitri, N, Gabbiani, C, Guerri, A. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
3O9B
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with kd25 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3O9L
 
 | | Design and optimisation of new piperidines as renin inhibitors | | Descriptor: | (3R,4S)-N-[2-chloro-5-(3-methoxypropyl)benzyl]-N-cyclopropyl-4-{4-[2-(2,6-dichloro-4-methylphenoxy)ethoxy]phenyl}piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Corminboeuf, O, Bezencon, O, Grisostomi, C, Remen, L, Richard-Bildstein, S, Bur, D, Prade, L, Hess, P, Strickner, P, Treiber, A. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and optimization of new piperidines as renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OAD
 
 | | Design and optimization of new piperidines as renin inhibitors | | Descriptor: | (3S,4R)-N-[2-chloro-5-(2-methoxyethyl)benzyl]-N-cyclopropyl-4-{6-[2-(2,6-dichloro-4-methylphenoxy)ethoxy]pyridin-3-yl}-4-hydroxypiperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Prade, L. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and optimization of new piperidines as renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NU6
 
 | |
