4NZP
 
 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
2AKO
 
 | |
4LXX
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Fuc3NFo and 5-N-Formyl-THF | | Descriptor: | (2R,3R,4S,5R,6R)-4-(formylamino)-3,5-dihydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | thoden, J.B, goneau, M.-F, gilbert, M, holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXY
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP and 10-N-Formyl-THF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXU
 
 | | dTdp-Fuc3N and 5-N-Formyl-THF | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXQ
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LY3
 
 | | Crystal structure of WlaRD, a sugar 3N-formyl transferase in the presence of dTPD-Qui3N, dTDP-Qui3NFo, and THF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, WlaRD a sugar 3N formyltransferase, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4LXT
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp-Qui3N and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4NOI
 
 | | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase subunit alpha, IODIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
6MQ8
 
 | | Binary structure of DNA polymerase eta in complex with templating hypoxanthine | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*IP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Hawkins, M.A, Jung, H, Lee, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 2020
|
|
4OC9
 
 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
5VLM
 
 | | Crystal structure of EilR in complex with crystal violet | | Descriptor: | CRYSTAL VIOLET, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
5VL9
 
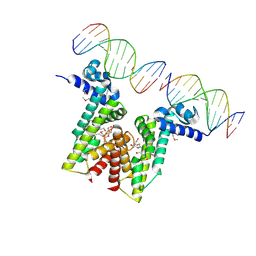 | | Crystal structure of EilR in complex with eilO DNA element | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*GP*TP*TP*GP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*CP*AP*AP*CP*TP*TP*TP*C)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
5VLG
 
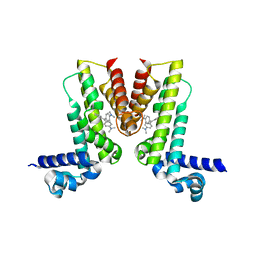 | | Crystal structure of EilR in complex with malachite green | | Descriptor: | MALACHITE GREEN, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.932 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3RFW
 
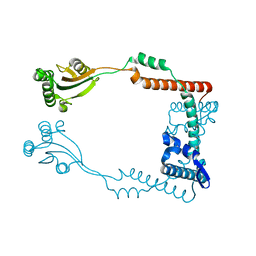 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Cell-binding factor 2 | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
3RGC
 
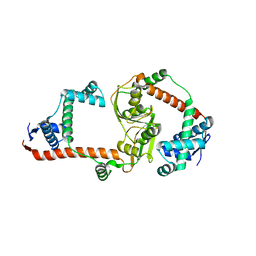 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Possible periplasmic protein | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
1BG7
 
 | | LOCALIZED UNFOLDING AT THE JUNCTION OF THREE FERRITIN SUBUNITS. A MECHANISM FOR IRON RELEASE? | | Descriptor: | CALCIUM ION, FERRITIN | | Authors: | Takagi, H, Shi, D, Ha, Y, Allewell, N.M, Theil, E.C. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Localized unfolding at the junction of three ferritin subunits. A mechanism for iron release?
J.Biol.Chem., 273, 1998
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
4NPX
 
 | | Structure of hypothetical protein Cj0539 from Campylobacter jejuni | | Descriptor: | Putative uncharacterized protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Adkins, J.N, Endres, M, Nissen, M, Konkel, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-11-22 | | Release date: | 2014-01-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of hypothetical protein Cj0539 from Campylobacter jejuni
To be Published
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | Descriptor: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3991 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
7C65
 
 | | Crystal structure of thioredoxin m1 | | Descriptor: | SODIUM ION, Thioredoxin M1, chloroplastic | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
